Weekly Recap (May 2025, part 2)
Comprehensive evolutionary modeling of human gene function, an AI reasoning model for rare disease diagnosis, an AI agent for scRNA-seq data exploration, FAIR scientific workflows, ...
This week’s recap highlights compendium of human gene functions derived from evolutionary modelling from the Gene Ontology Consortium, an AI reasoning model applied to rare disease diagnosis, an agentic AI for scRNA-seq data exploration, and applying FAIR principles to scientific workflows.
Others that caught my attention include polars-bio for fast operations on genomic intervals, LiftOn for combining DNA and protein alignments to improve genome annotation, a review on modeling and design of transcriptional enhancers, cell type annotations with ChatGPT, single-molecule protein sequencing with nanopores, genomic language models for predicting enhancers, a new cell segmentation method for spatial transcriptomics, domain-specific embeddings to uncover latent genetics knowledge, demographic inference from genealogical trees, a de novo metagenome assembler for heavily damaged ancient datasets, and an R package for epigenome-wide association studies.
Deep dive
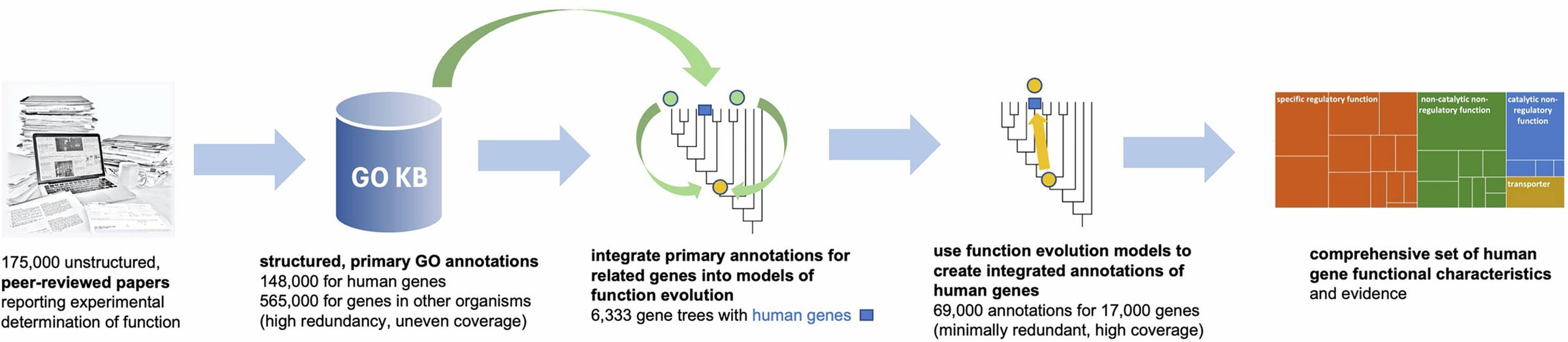
A compendium of human gene functions derived from evolutionary modelling
Paper: Feuermann, et al, “A compendium of human gene functions derived from evolutionary modelling,” Nature, 2025, https://doi.org/10.1038/s41586-025-08592-0.
The Gene Ontology (geneontology.org) was started in the 1990s and provides what’s probably the largest collection of functional annotation information about genes and gene products. There are over 43,000 hits in PubMed for Gene Ontology, and scores of software tools for analyzing genomic data with GO, from enrichment analysis to functional annotation and many others.
TLDR: This paper comprehensively maps human gene functions using evolutionary modeling, enhancing our understanding of genome biology and functional genomics. The authors integrate experimental gene function data from humans and model organisms to create detailed evolutionary models, capturing functional gains and losses, providing an enriched resource that substantially improves current functional annotations. This integrated dataset notably improves common genomic analyses, like Gene Ontology enrichment.
Summary: Feuermann and colleagues provide an extensive and integrative resource of human gene functions, developed by evolutionary modeling of functional annotations derived from experimental data. By leveraging annotations from model organisms, the authors created evolutionary models across gene families, systematically documenting functional gain and loss events, resulting in a rich dataset that includes functional annotations for approximately 82% of human protein-coding genes. The importance of this work lies in addressing gaps left by previous databases, which lacked integration of experimental evidence across species and evolutionary contexts, leading to a resource that is both comprehensive and highly reliable. This dataset enhances the accuracy and utility of common bioinformatics approaches, particularly Gene Ontology enrichment analyses, by offering non-redundant and experimentally validated annotations. Applications of this work include more accurate gene function prediction, improved interpretation of genomic data, and insights into the evolutionary dynamics of gene functions.
Methodological highlights:
Implements explicit evolutionary models to systematically reconstruct the historical gain and loss of gene functions across human protein-coding genes.
Selectively integrates experimental annotations from extensive literature, prioritizing minimally redundant, maximally informative GO terms.
Provides reliability estimates demonstrating high confidence (90–97%) in these evolutionary-based functional annotations.
New tools, data, and resources:
PAN-GO dataset: A publicly available, comprehensive set of integrated GO annotations based on evolutionary modeling for human genes, accessible at https://functionome.geneontology.org.
PAINT tool: Software for manual curation and integration of phylogenetic data with GO annotations, allowing users to explore and contribute to gene function evolutionary models. Available at https://github.com/pantherdb/db-PAINT.
Evolutionary model browser: https://pantree.org.
Evidence Aggregator: AI reasoning applied to rare disease diagnostics
Paper: Twede et al., “Evidence Aggregator: AI reasoning applied to rare disease diagnostics,” bioRxiv, 2025, https://doi.org/10.1101/2025.03.10.642480.
I spent my grad school and postdoc years in the late 2000s in genetic epidemiology, right around when the first GWAS were being performed on AMD. I wish a tool like this (and data to power it) existed back then.
TLDR: This paper introduces an AI-driven pipeline that greatly reduces the burden of manual literature review in rare disease diagnosis. The Evidence Aggregator (EvAgg) automates extraction and synthesis of literature-based genetic variant evidence, significantly speeding up the diagnostic workflow and demonstrating strong benchmark performance, including a notable 34% reduction in analyst review time.
Summary: The authors describe Evidence Aggregator (EvAgg), an AI pipeline leveraging large language models (LLMs) to automate literature review for diagnosing rare genetic diseases. EvAgg consists of three modules: selecting relevant papers, identifying genetic variants from these papers, and extracting detailed content such as phenotype, variant type, and inheritance patterns. The significance of this work lies in addressing the major bottleneck of manually reviewing and synthesizing genetic evidence from extensive literature, thereby expediting rare disease diagnoses and potentially increasing diagnostic accuracy and efficiency. Practical applications include facilitating faster variant assessments, supporting genetic analysts in clinical and research settings, and potentially reducing diagnostic latency for patients suffering from rare diseases.
Methodological highlights:
Implements a consensus-based approach combining keyword matching and LLM classification to select relevant literature with extremely high recall (97%).
Employs normalization and validation techniques, including HGVS nomenclature standards and external tools such as Mutalyzer, to robustly extract genetic variant data.
Includes rigorous benchmarking against manually curated datasets and incorporates user feedback to evaluate real-world performance and utility in clinical workflows.
New tools, data, and resources:
Evidence Aggregator (EvAgg): An open-source AI tool designed to automate literature review for rare genetic disease diagnostics, significantly reducing review time and enhancing variant analysis workflows. Available at https://github.com/microsoft/healthfutures-evagg, implemented primarily using Python and Azure OpenAI services.
Integrated seqr analysis interface: EvAgg outputs have been integrated into seqr, a web-based genome analysis platform (https://seqr.broadinstitute.org/), facilitating direct incorporation into existing genomic diagnostic workflows.

CompBioAgent: An LLM-powered agent for single-cell RNA-seq data exploration
Paper: Zhang, et al., “CompBioAgent: An LLM-powered agent for single-cell RNA-seq data exploration,” bioRxiv, 2025, https://doi.org/10.1101/2025.03.17.643771.
TLDR: This paper introduces a chatbot-like interface for single-cell RNA-seq analysis that converts plain-language queries into detailed data visualizations. CompBioAgent integrates LLMs with databases and visualization tools to automate single-cell data exploration, lowering barriers for non-computational researchers.
Summary: Zhang and colleagues present CompBioAgent, a web-based AI tool leveraging large language models (LLMs) to streamline the analysis and visualization of single-cell RNA sequencing data without requiring coding expertise. Users interact with CompBioAgent via natural language queries, which the system then translates into structured database requests and visualization commands. This tool addresses a significant bottleneck by automating complex bioinformatics workflows, making high-dimensional single-cell data analyses accessible to a broader audience. CompBioAgent notably integrates with CellDepot for dataset access and Cellxgene for visualization, providing rich graphical outputs such as UMAP embeddings, heatmaps, and violin plots, facilitating quicker insights into disease mechanisms and cellular behaviors.
Methodological highlights:
Uses prompt engineering with both simple and complex structured templates to effectively convert user queries into precise JSON-formatted commands.
Employs few-shot prompting techniques to enhance the accuracy and flexibility of interpreting user input through various LLMs.
Implements robust error detection and correction to handle inconsistent outputs from LLMs, thus ensuring stable and reliable user interactions.
New tools, data, and resources:
CompBioAgent: A web app using natural language to query and visualize scRNA-seq datasets, primarily implemented using OpenAI's GPT, Anthropic Claude, Groq, DeepSeek R1, and local Llama 3 models; source code at https://github.com/interactivereport/compbioagent.
CellDepot Integration: Comprehensive single-cell RNA-seq database used by CompBioAgent for data querying; available at https://celldepot.bxgenomics.com/compbioagent.
Cellxgene VIP: An advanced web platform integrated into CompBioAgent for high-resolution, interactive single-cell visualizations including embedding plots, heatmaps, and dot plots; detailed at https://github.com/interactivereport/cellxgene_VIP.

Applying the FAIR Principles to computational workflows
Paper: Wilkinson, et al., “Applying the FAIR Principles to computational workflows,” Scientific Data, 2025, https://doi.org/10.1038/s41597-025-04451-9.
You all know by now I’m a Nextflow evangelist. Search this blog at the top for “Nextflow” and you’ll find coverage of dozens of papers either developing tools in Nextflow or applying Nextflow pipelines for analysis. I’m also a fan of what Seqera and the nf-core community are doing for the life sciences community. This paper is about FAIR principles in computational workflows — something that’s baked into the very core of Nextflow and the nf-core ecosystem.
TLDR: This paper caught my eye because it thoughtfully extends the FAIR (Findable, Accessible, Interoperable, Reusable) principles to computational workflows, significantly clarifying how researchers can practically enhance reproducibility and usability in computational research. The authors systematically adapt and apply FAIR principles for both software and data to computational workflows, providing concrete guidelines and illustrative examples to improve workflow discoverability, interoperability, and reusability.
Summary: Wilkinson and colleagues present a detailed discussion and practical recommendations for applying the FAIR principles (originally designed for data and software) to computational workflows. The FAIR workflows working group (WCI-FW) has addressed this by carefully considering how workflows, as complex combinations of software and data, can be structured to improve their FAIRness. The significance of this contribution lies in its potential to enhance scientific reproducibility and collaborative sharing by making workflows easier to find, access, integrate, and reuse across different research environments. Methodologically, the paper provides explicit recommendations, such as assigning persistent identifiers to workflow versions, using standardized metadata schemas (like CWL and RO-Crate), and ensuring clear licensing and provenance documentation. These recommendations have wide-ranging implications for computational biology and other fields reliant on reproducible computational analysis, facilitating more robust and transparent science.
Methodological highlights:
Clearly defines workflow-specific interpretations of FAIR principles, accounting for workflows’ composite nature, including their executable and data components.
Proposes detailed metadata standards (e.g., CWL, RO-Crate, schema.org) and encourages persistent identifiers for individual workflow components, enhancing their discoverability and traceability.
Highlights practical considerations for ensuring long-term accessibility and reproducibility, especially regarding software dependencies and execution environments.
New tools, data, and resources:
WorkflowHub (previously covered here): a registry for workflows providing unique DOIs and rich metadata to enhance workflow discoverability and reuse (https://workflowhub.eu).
Dockstore: a platform for sharing reproducible and accessible computational workflows, supporting multiple workflow languages (dockstore.org).
RO-Crate and CWLProv: standards for packaging and documenting workflows and their provenance, making computational workflows more transparent and easier to reuse and interpret (https://www.researchobject.org/workflow-run-crate/profiles/0.1/provenance_run_crate).

Other papers of note
polars-bio - fast, scalable and out-of-core operations on large genomic interval datasets https://www.biorxiv.org/content/10.1101/2025.03.21.644629v1
Combining DNA and protein alignments to improve genome annotation with LiftOn https://genome.cshlp.org/content/35/2/311
Review: Modelling and design of transcriptional enhancers https://www.nature.com/articles/s44222-025-00280-y
Beyond the Hype: The Complexity of Automated Cell Type Annotations with GPT-4 https://www.biorxiv.org/content/10.1101/2025.02.11.637659v2
Toward single-molecule protein sequencing using nanopores https://www.nature.com/articles/s41587-025-02587-y (read free: https://rdcu.be/edWPi)
Genomic Language Model for Predicting Enhancers and Their Allele-Specific Activity in the Human Genome https://www.biorxiv.org/content/10.1101/2025.03.18.644040v1
Segger: Fast and accurate cell segmentation of imaging-based spatial transcriptomics data https://www.biorxiv.org/content/10.1101/2025.03.14.643160v1
Domain-specific embeddings uncover latent genetics knowledge https://www.biorxiv.org/content/10.1101/2025.03.17.643817v1
A likelihood-based framework for demographic inference from genealogical trees https://www.nature.com/articles/s41588-025-02129-x (read free: https://rdcu.be/eejtr)
CarpeDeam: A De Novo Metagenome Assembler for Heavily Damaged Ancient Datasets https://www.biorxiv.org/content/10.1101/2024.08.09.607291v2
easyEWAS: a flexible and user-friendly R package for epigenome-wide association study academic.oup.com/bioinformaticsadvances/article/5/1/vbaf026/8011369

