Weekly Recap (January 2, 2026)
NSF reorg, Science in 2050, 2025 LLM recap, R+Python, R Data Scientist, virtual cells, genomics in 2026, Claude Code course, AI and labor, how uv got so fast, Anthropic/biotech, papers+preprints
Science: The National Science Foundation just had a big reorganization. Here are five things to know.
Nature: Science in 2050: the future breakthroughs that will shape our world.
Simon Willison: 2025: The year in LLMs.
Boaz Barak at Less Wrong: You will be OK. Barak argues that while society should take low-probability AI catastrophes seriously, individuals are better served by focusing on what they can control, expecting AI-driven change to be fast but uneven and gradual rather than a single “AGI moment,” and staying engaged, optimistic, and psychologically steady without becoming complacent.
Emily Riederer: R + Python: From polyglot to crosspolination.
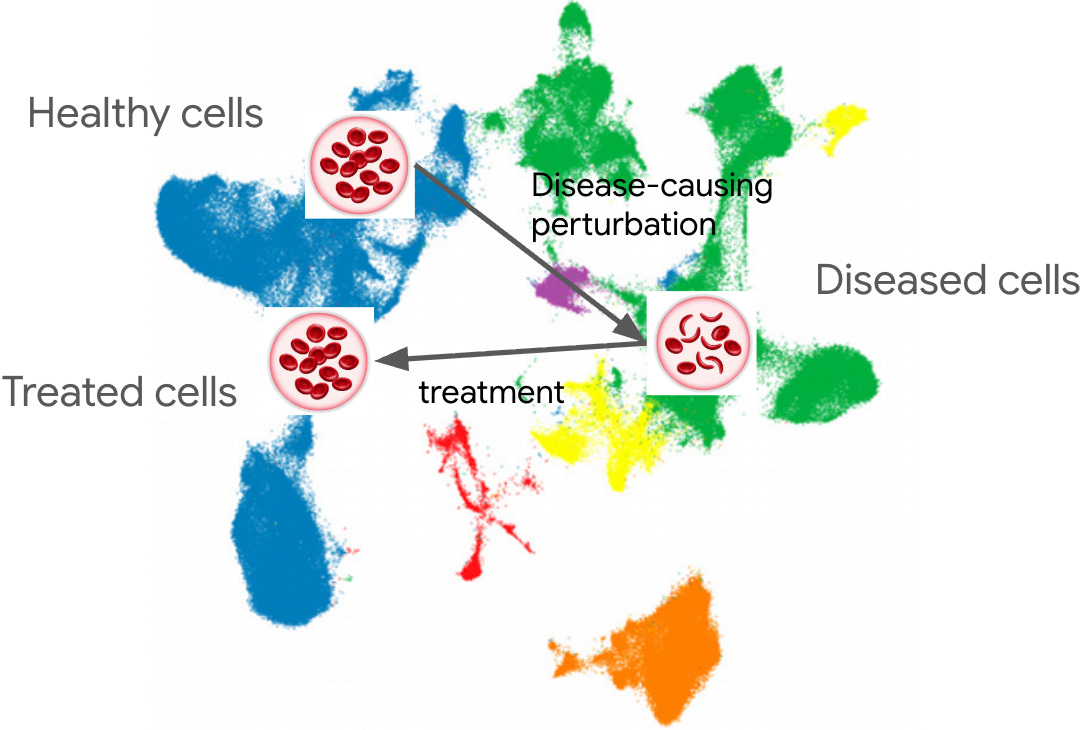
Andrew Carroll: The Virtual Cell Will Be More Like Gwas Than Alphafold.
Genomics in 2026: Trends and Predictions from the Fulcrum Genomics Team: AI in Genomics: Powerful, but Not a Magic Wand; Standards Cross the Academia–Industry Divide; The Genomics Stack Grows Up: Tools, Workflows, and Rust; From Short Reads to Structural Truth: Long Reads, SVs, and Error-Corrected Sequencing; Beyond the Genome: Proteins Take Center Stage; Looking Ahead: Making Genomics Work in the Real World.
Anthropic launches a free short course on using Claude Code. In the course you’ll:
Understand the underlying architecture of Claude Code, the tools it uses to navigate your codebase, and how it stores memory across sessions.
Explore and understand the codebase of a RAG chatbot and how information flows between the frontend and the backend.
Initiate a CLAUDE.md file inside your project directory containing information and guidelines about your codebase that Claude Code can remember across sessions.
Get context into Claude Code by mentioning the relevant files and providing screenshots or images, and control the context using escape, clear, and compact commands.
Add features to the frontend and backend of the RAG chatbot: ask Claude Code to plan first to improve its performance, use thinking mode for harder tasks, and brainstorm ideas using Claude Code’s subagents.
Write tests to evaluate the RAG chatbot functionalities, and refactor parts of the chatbot.
Use git worktrees to run multiple Claude sessions simultaneously, each focused on adding an independent feature to the chatbot.
Fix Github issues, and create, review and merge Github pull requests using Claude Code’s Github integration.
Execute code before and after using tools through Claude Code hooks.
Refactor a Jupyter notebook for e-commerce data analysis and transform it into a dashboard.
Connect Claude to the Figma MCP server to import a design mockup to Claude Code, and develop a web interface showing economic data from the Federal Reserve Economic Data.
Use Playwright MCP server to automatically open a web browser, take screenshots, and guide Claude Code to improve the UI design of an application.
Sal Khan: A.I. Will Displace Workers at a Scale Many Don’t Realize.
The R Data Scientist 2025-12-30: R ecosystem radar, R package engineering, hands-on R analysis, stats & inference, academic research.
Andrew Nesbitt: How uv got so fast. (See also my series on uv, part 1, 2, 3, 4).
When A.I. Took My Job, I Bought a Chain Saw.
Is Anthropic becoming a biotech company? They’re advertising life sciences positions.
Finally, a few other papers and preprints that caught my attention this week:
A SNP panel for co-analysis of capture and shotgun ancient DNA data
R2G2: A Python-R Framework for Seamless Integration of R/Bioconductor Tools into Galaxy
(Nucleotide Transformer v3) A foundational model for joint sequence-function multi-species modeling at scale for long-range genomic prediction (pdf)
ColabPCR: A validated Google Colaboratory Notebook for Reproducible and Precise Primer Design
CRISPR-HAWK: Haplotype- and Variant-aware guide design toolkit for CRISPR-Cas
Clair3-RNA: a deep learning-based small variant caller for long-read RNA sequencing data
Creating Enforceable Biosecurity Standards for Nucleic Acid Providers
Paleogenomic insight into the collapse, recovery, and management of American bison
Related: my post from yesterday on how I find blogs, news stories, and papers like what’s in this post.
Staying Current in Data Science and Computational Biology: 2026 Edition
I started blogging 17 years ago, and the most common question I get from colleagues and students is how do you stay current and find all these interesting papers/blogs/etc.? I wrote this post in 2012 and followed it up 5 years later with this post in 2017





The bit about virtual cells being more like GWAS than AlphaFold is underappreciated. When you're working with transcriptomic perturbation data, the relationship between disease state and treatment isnt a clean foldable protein structure problem its noisy, probabilistic, and heavily context-dependent. The expectation that we'd just get another AlphaFold moment for cells misses how fundamentaly different the data challenge is when you're dealing with complex systems rather than single-molecule structures.