Closing my tabs (Sep 5 2025)
NIH budget, AI, R, Bluesky & Science, state of biotech, AI in higher ed, R packages Top 40, structural variation, statistical power, Python documentary, Manhattan Project, tidymodels, ...
Happy Friday, colleagues. Somehow it’s September (I did not approve of this). Lots going on this week, and this is my regular attempt to close out my browser tabs I’ve accumulated over the past week with blog posts, podcasts, papers, etc. in AI, data science, genomics, public health, programming, scicomm, and other miscellany. Enjoy!
A glimmer of good news in dark times: House Republicans add to support for maintaining NIH budget in 2026. Both chambers of Congress have now rejected Trump’s proposed 40% cut.
Posit’s AI Newsletter, first edition. This started out as an internal update at Posit. It’ll be published every other week recapping what’s happening at Posit and in the broader world of AI in data science.
A new paper from Heng Li: Finding easy regions for short-read variant calling from pangenome data.
R Weekly 2025-W36 newsletter: Creating messy datasets for teaching purposes with {truffle}, 25 things you didn’t know you could do with R, OpenAI Codex in Positron.
Nature: Research posts on Bluesky are more original — and get better engagement.
Anyone can be the best: Impact of diverse methodologies on the evaluation of structural variant callers. In a previous position I thought a lot about structural variation, and I’ve reviewed lots of new SV methodology in previous recap posts here. This new preprint shows lots of variation in SV discovery/calling tools depending on the choice of ground truth, reference genome, and genomic regions used for evaluation.
July 2025 Top 40 New CRAN Packages. Joe Rickert’s picks for the best new packages to land on CRAN in July, organized in thirteen categories: Causal Inference, Computational Methods, Data, Ecology, Epidemiology, Machine Learning, Mathematics, Medical Statistics, Pharma, Statistics, Time Series, Utilities, and Visualization.
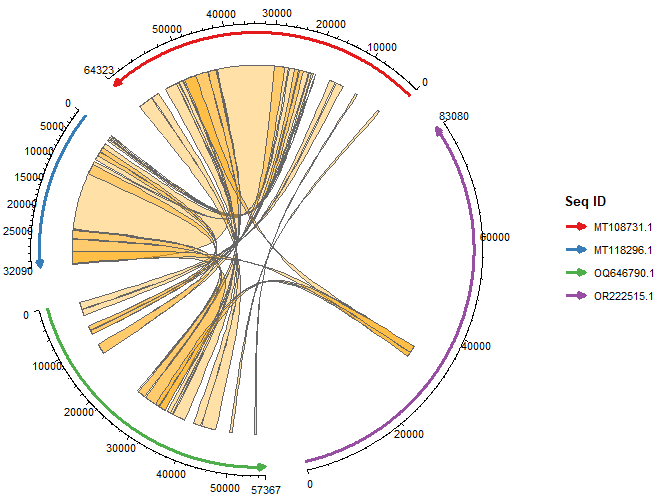
(From the post above): The ggchord R package extends ggplot2 to visualize pairwise BLAST alignment results as chord diagrams, intuitively displaying homologous regions between query and subject sequences. See the vignette.
Simon Couch: I was wrong about tidymodels and LLMs. Modern frontier models “just know” the tidymodels ecosystem much better than they did years ago.
On STAT: New report details the state of the biotech industry in Massachusetts. The results are grim. Venture capital investment in Massachusetts-based companies dropped more than 17% to $2.75 billion in the first half of 2025, compared to the same period in 2024, the lowest level since 2017.
Ashlee Vance hosted Cathy Tie on the Core Memory podcast: Cathy Tie Is Ready To Gene Edit Babies. Cathy and Eriona have put together a well-written ethics statement on the work they’re planning at Manhattan Genomics.
Nature: Hundreds of suspicious journals flagged by AI screening tool.
Science: AI enters the grant game, picking winners. Funders test algorithms to spot promising science, raising hopes of faster reviews and fears of bias.
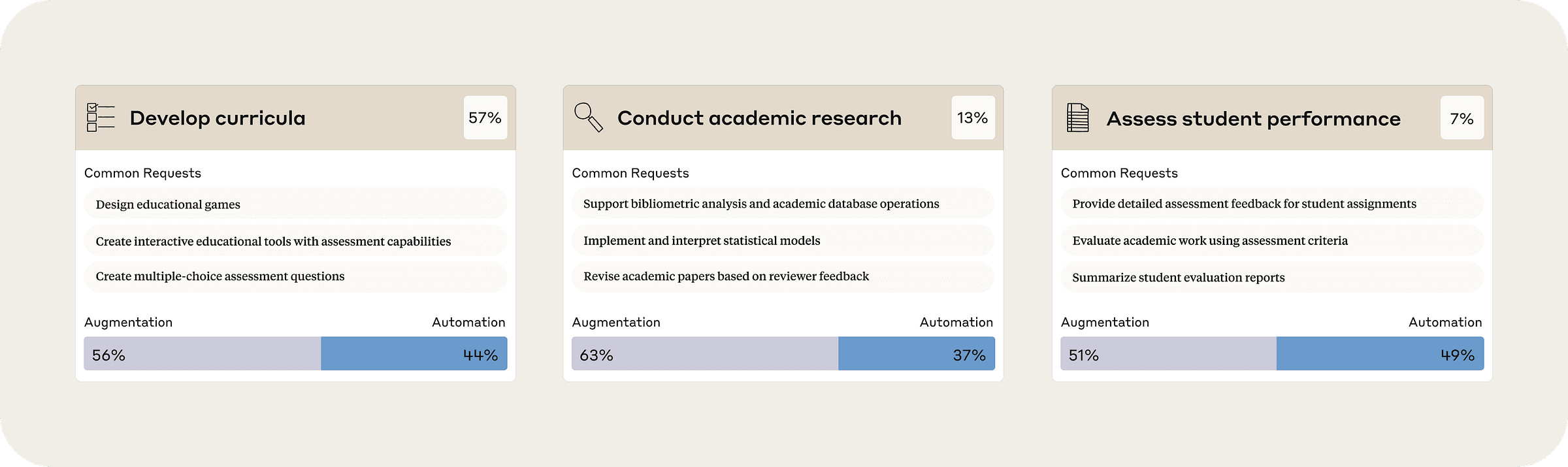
Anthropic Education Report: How educators use Claude. Anthropic analyzed 74,000 anonymized conversations to understand how university educators are using AI. 7% of higher ed instructors' AI chats in its sample involved developing curricula, 13% were conducting academic research, and 7% involved assessing students' performance. Anthropic says educators used AI for grading less often than other tasks. But 48.9% of the Claude conversations about grading turned the task fully over to the bot in ways that researchers found “concerning.”
Python: The Documentary | An origin story. What began as a side project in Amsterdam during the 1990s became the software powering AI, data science, and some of the world’s biggest companies. But Python's future wasn't certain; at one point it almost disappeared. This 90-minute documentary features Guido van Rossum, Travis Oliphant, Barry Warsaw, and many more, and they tell the story of Python’s rise, its community-driven evolution, the conflicts that almost tore it apart, and the language’s impact on... well… everything.
Claus Wilke: PhD-level abilities and character traits. Lots of talk out there about “PhD-level intelligence” with the latest iteration of frontier AI models, the latest being Sam Altman suggesting GPT-5 is like a “team of Ph.D. level experts in your pocket.” Getting a PhD is not just about being “smart.” As Claus Wilke explains in this post, success in a PhD program depends on traits like executive function, fearlessness, resilience, discipline, motivation, curiosity, empathy, and a strong sense of ethics, which all matter more than raw intelligence (however that’s measured).
Carlisle Rainey: A One-Page Primer on Statistical Power. Statistical power is the chance to reject the null when it’s false. Why it matters, how to compute it, and why both researchers and readers should care. This is a one-page primer with rules of thumb and key readings.
Humans are being used to fix AI slop. The widespread adoption of AI for everything has created a new type of work: fixing AI’s mistakes. Designers are being hired to remake sloppy AI art. Actual human writers are hired to make ChatGPT’s writing sound more human. If you’ve vibe-coded yourself into oblivion, my contact information is easy to find but I’m not cheap.
nf-core blog: Running nf-core pipelines in Google Colab.
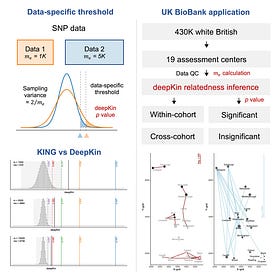
And finally, a shameless plug: earlier this week I posted a recap with a deep dive on some papers I’ve been catching up on: Biobank-scale relatedness estimation, SNP calling and phasing with long RNA-seq reads, predicting expression-altering promoter mutations with deep learning, cross-species filtering for comparative genomics, and a few others.
Weekly Recap (Sep 2025 part 1)
This week’s recap highlights biobank-scale relatedness estimation, SNP calling and haplotype phasing with long RNA-seq reads, predicting expression-altering promoter mutations with deep learning, and cross-species filtering for reducing alignment bias in comparative genomics studies.