Weekly recap (Oct 17, 2025)
Bioconductor on GPUs, tidyomics, Quarto 1.8, Anthropic red-teaming on AIxBio, PGS in the clinic, Shock Doctrine in genome engineering, GenAI for data viz with R, Datapalooza, 4 compact rejections
GPU Support in Bioconductor. GPU acceleration enables faster and more scalable analysis workflows, especially for tasks like deep learning, image processing, and large-scale genomics. Bioconductor is developing better support for maintainers authoring GPU-capable packages, including adding a new Nvidia GPU build machine, new release and devel (Nvidia) GPU software builds, GPU-aware containers, and a biocViews GPU term. This post shares these new resources and how Bioconductor package maintainers can take advantage of them.
The agenda for Datapalooza 2025 conference at the University of Virginia School of Data Science has been released. The theme is Truth and Accountability in the Age of AI. You can register here (free).
Penn and USC reject the compact.
Quarto 1.8 released: New features include integrated Axe-core checks for HTML documents, access to environment variables via code cells, light/dark mode logos and colors, and Brand Extensions for sharing your org’s brand.
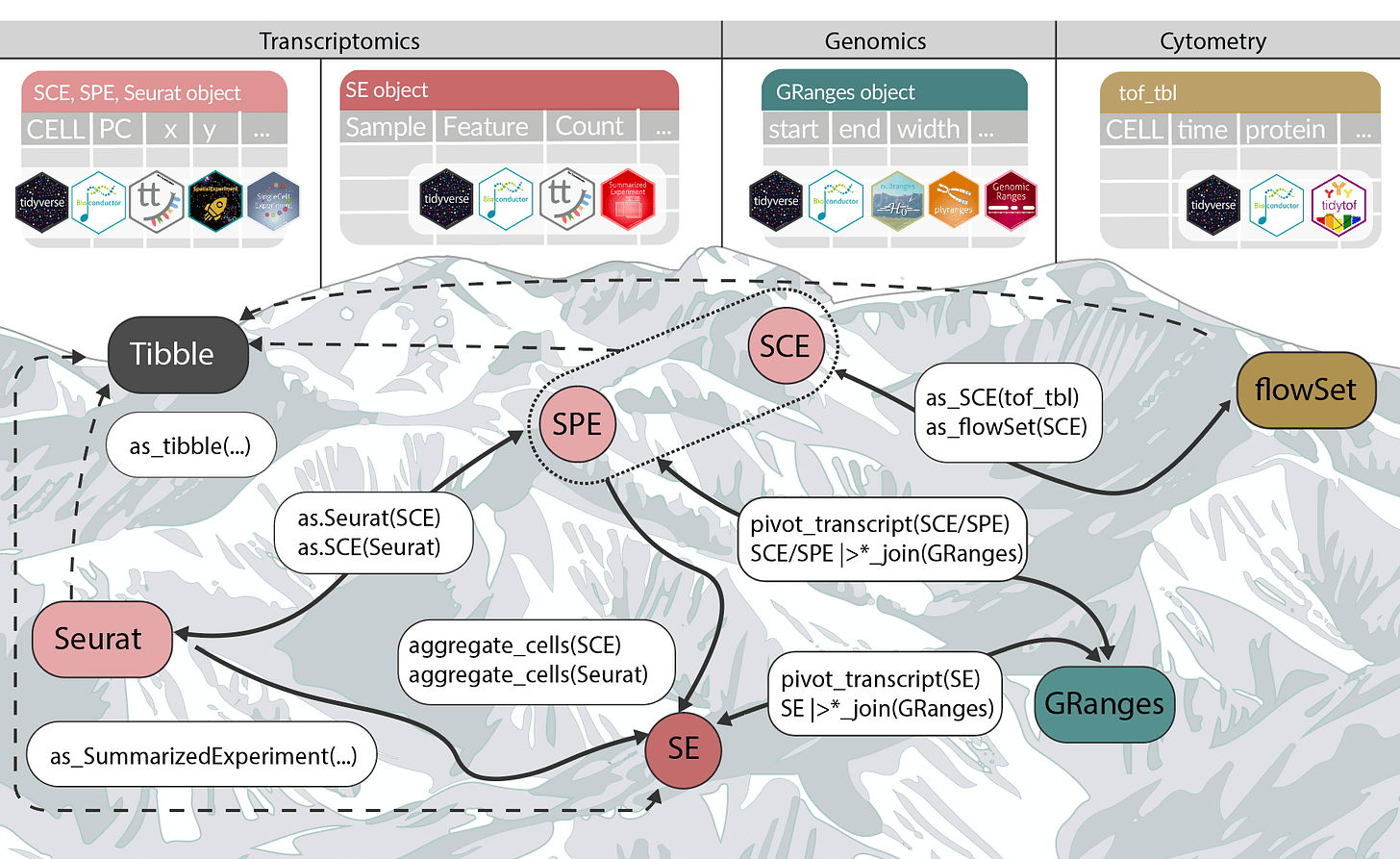
Tidyomics blog: The Tidyomics Ecosystem. An introduction to the tidyomics ecosystem, including principles, core packages, publications, and community resources for tidy omic analysis. Tidyomics is an open project to develop and integrate software and documentation to enable a tidy data analysis framework for omics data objects (Hutchison et al. 2024).
R Weekly 2025-W42: mirai, genAI for data visualization, behavior-driven development, much more.
In that GenAI for data visualization post, Nicola Rennie asks: Can generative AI create good data visualisations? This blog post compares the performance of ChatGPT, Claude, Copilot, and Gemini when presented with a generic request to visualise a dataset, and offers a useful recap (Gemini best for now).
Gemini: Good overall with comparable performance across the two datasets and prompts. The charts aren’t perfect, but they allow interactive data exploration and don’t over-complicate it.
ChatGPT: Reasonable overall with comparable performance across the two datasets and prompts. The charts look more amateurish and poorer quality compared to Gemini.
Claude: Good for small datasets but does way too much, unless you prompt it not to. Unable to work with larger data.
Copilot: Poor overall. Produced code to create a chart, but not the chart itself. Unable to work with larger data.
Blaze Newsletter: Data Scientist with R: Academic Research, Statistical Inference, Shiny Engineering, R + AI Apps, R Visualisation, R Community News.
Newsletter: red.anthropic.com: research from Anthropic’s Frontier Red Team on what frontier AI models mean for national security. Sep 2025 article: Why do we take LLMs seriously as a potential source of biorisk?
What can you do with Positron Assistant? I wrote about Positron Assistant a few months ago. This very short video provides a quick look at the highlights.
STAT1: In biotech, Boston reigns supreme, but its competitive edge is being challenged. Federal funding cuts and threats to higher education are undermining the city’s recipe for success.
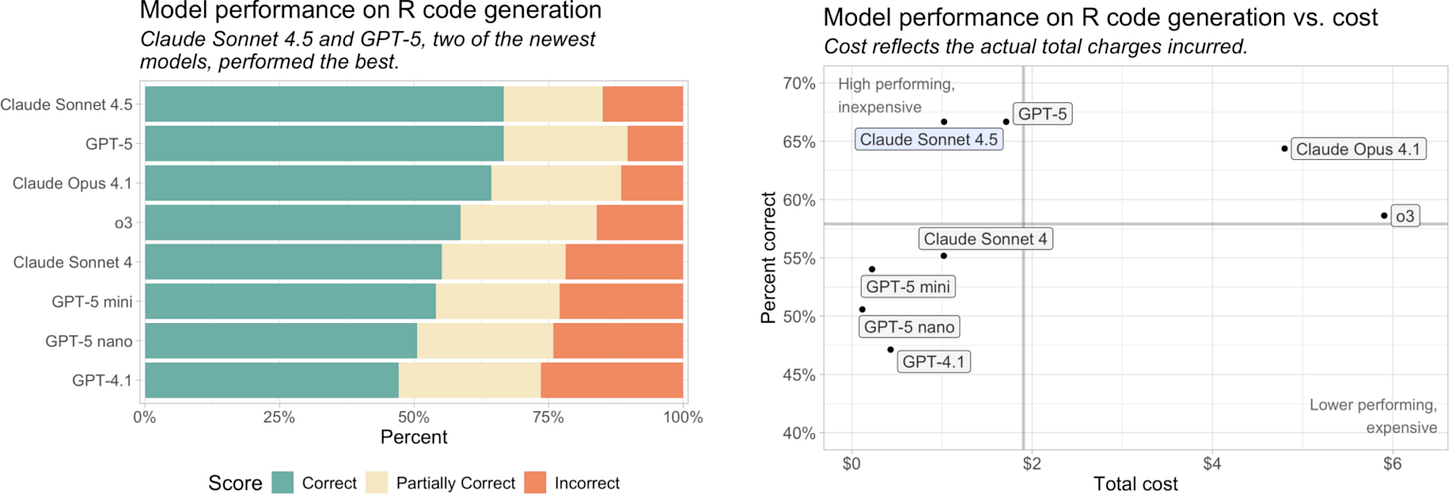
Posit blog: 2025-10-10 AI Newsletter. Claude Sonnet 4.5, Investments in and from OpenAI, Posit News. In evaluating R coding capabilities, Sonnet 4.5 tied with GPT-5 in percentage correct while being slightly cheaper.
Yet Another AI Bubble Article (YAABA): NYT - The A.I. Bubble Looks Real.
U.S. Biotech Future Is Now Made in China.
Nature Human Behaviour paper: How to develop good research questions.
Isabella Velásquez on the Posit Blog: posit::conf(2025) recap.
Docker blog: How to Do Hardened Images (and Container Security) Right.
Chris Loy: The AI coding trap.
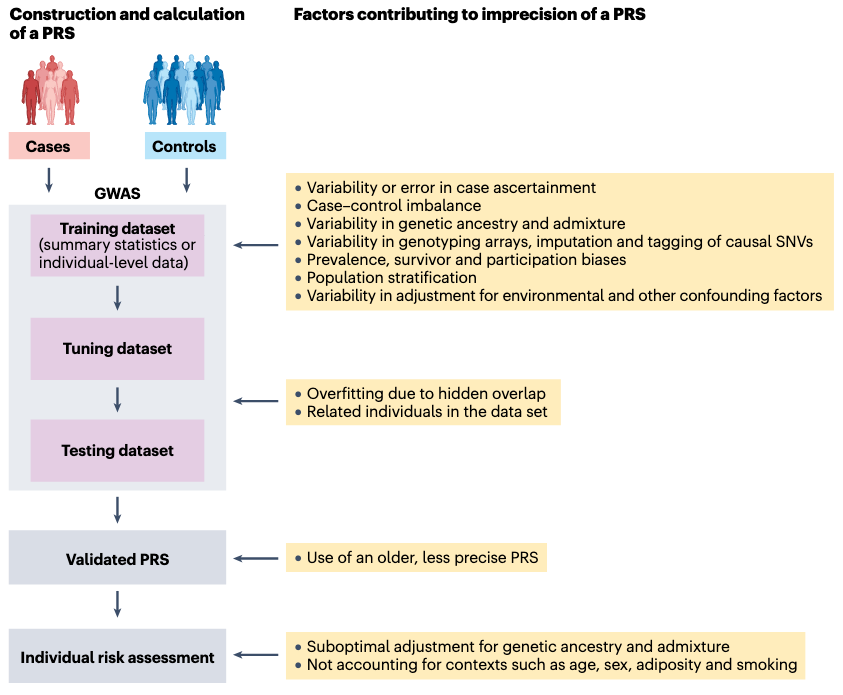
Nature Reviews Genetics: Clinical use of polygenic risk scores. This is an incredibly comprehensive review on polygenic scores, with nearly 200 references and related links.
The International Union for Conservation of Nature (IUCN) is weighing a proposed ban on unleashing genetically modified plants or animals into the wild. I’m highly opinionated here,2 and in almost all cases I think broad moratoria are a bad substitutes for careful, measured, and well-regulated progress (see a recent essay from my friend and colleague, Alexander Titus: Shock Doctrine in the Life Sciences - When Fear Overwhelms Facts).
Washington Post: Scientists say they revived an extinct wolf species. A vote will decide their future.
Nature: Should genetically modified wildlife be banned? Scientists weigh the risks
Nathan Benaich’s State of AI report. You can catch the highlight in Tweet thread or YouTube.
Finally, a few other papers and preprints that caught my attention this week:
Accurate somatic small variant discovery for multiple sequencing technologies with DeepSomatic
Efficient and accurate search in petabase-scale sequence repositories
Nextpie: A web-based reporting tool and database for reproducible Nextflow pipelines
The utility of ultra-deep RNA sequencing in Mendelian disorder diagnostics
BioRAGent: natural language biomedical querying with retrieval-augmented multiagent systems
From genotype to phenotype with 1,086 near telomere-to-telomere yeast genomes
Lesson planning with ChatGPT for inquiry-based biology instruction – A(I) roll of the dice?
A Tribal data repository to advance Indigenous health and sovereignty
METAHIT enables comprehensive and flexible genome-resolved microbiome analysis with metagenomic Hi-C
If you’re at the University of Virginia, the library provides free access to all STAT+ Articles. Register through the proxy here, as described on the HSL guide. If you’re at another university, especially an R1 research university with a medical school, check your library or health sciences library. You probably have access and don’t know about it.
I’m not an unbiased observer here. I worked at Colossal Biosciences for a few years before coming back to UVA. In the past few months I’ve published two review papers on how genome engineering can provide benefits to biodiversity conservation: (1) In Nature Reviews Biodiversity: Genome engineering in biodiversity conservation and restoration (blog post); (2) In Journal of Heredity: De-extinction technology and its application to conservation.