Weekly Recap (Nov 7, 2025)
LLMs for learning to code, R updates (R Data Scientist, R Weekly), RAG, layoffs & AI, DuckDB for data science, Anthropic+Iceland, AI in science, AI in drug discovery, Nextflow & pathogen surveillance
Andrew Heiss: LLMs are bad and dangerous for learning how to code in this class (Data Visualization with R).
The R Data Scientist November 4 2025: R ecosystem updates, workflows & techniques, ggplot2 visuals, mapping with R, Shiny testing and QA, academic research.
R Weekly 2025-W45: Pledging my time, neon ghosts, Bioconductor release.
Steven P. Sanderson: RAG with Ollama and ragnar in R: A Practical Guide for R Programmers. Learn how to build a privacy-preserving RAG workflow in R using Ollama and the ragnar package. Discover step-by-step methods for summarizing health insurance policy documents, automating compliance reporting, and leveraging local LLMs—all within your R environment. I wrote something similar a few months ago (“Tidy RAG in R with ragnar”) but Steve’s tutorial is much better than mine.
Claus Wilke: LLMs excel at programming—how can they be so bad at it? “My explanation for the mystery of why LLMs can be both exceptionally good and quite terrible at programming.”
FT: Layoffs and AI.
Khuyen Tran: A Deep Dive into DuckDB for Data Scientists. If you want to follow along, here is the Marimo notebook with the code and output.
Anthropic and Iceland announce a national AI education pilot.
Nature: PhD training needs a reboot in an AI world.
Nature: Dismantling of US federal agencies will ‘destroy science’
Science: Letters to scientific journals surge as ‘prolific debutante’ authors likely use AI.
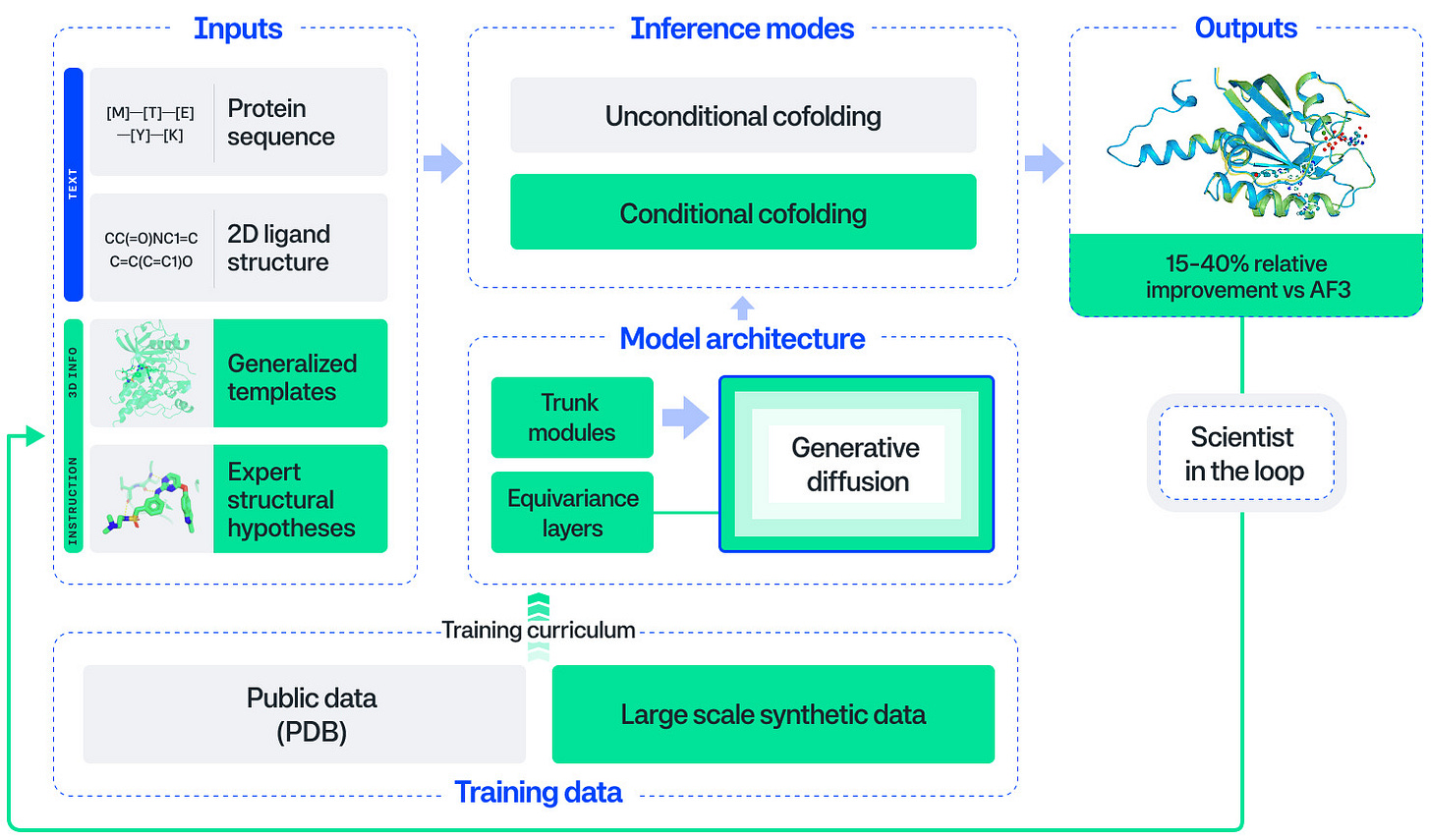
Pearl: The Next Generation Foundation Model for Drug Discovery. See also, the preprint: Pearl: A Foundation Model for Placing Every Atom in the Right Location.
NVIDIA blog: Introducing the CodonFM Open Model for RNA Design and Analysis. See also the manuscript: Learning the Language of Codon Translation with CodonFM.
Ten essential tips for robust statistics in cell biology. Spoiler alert:
Design your study with statistics in mind
Understand your data structure and sources of noise
Explore the data before statistical testing
Perform correction for multiple-hypothesis testing
Emphasize effect sizes and confidence intervals over P values
Validate findings in truly independent datasets
Account for biological and technical variation
Distinguish between correlation and causation
Make your analysis reproducible and transparent
Collaborate early with biostatisticians
R-Ladies STL recording: Efficient File Management in R with {fs} with Jadey Ryan.
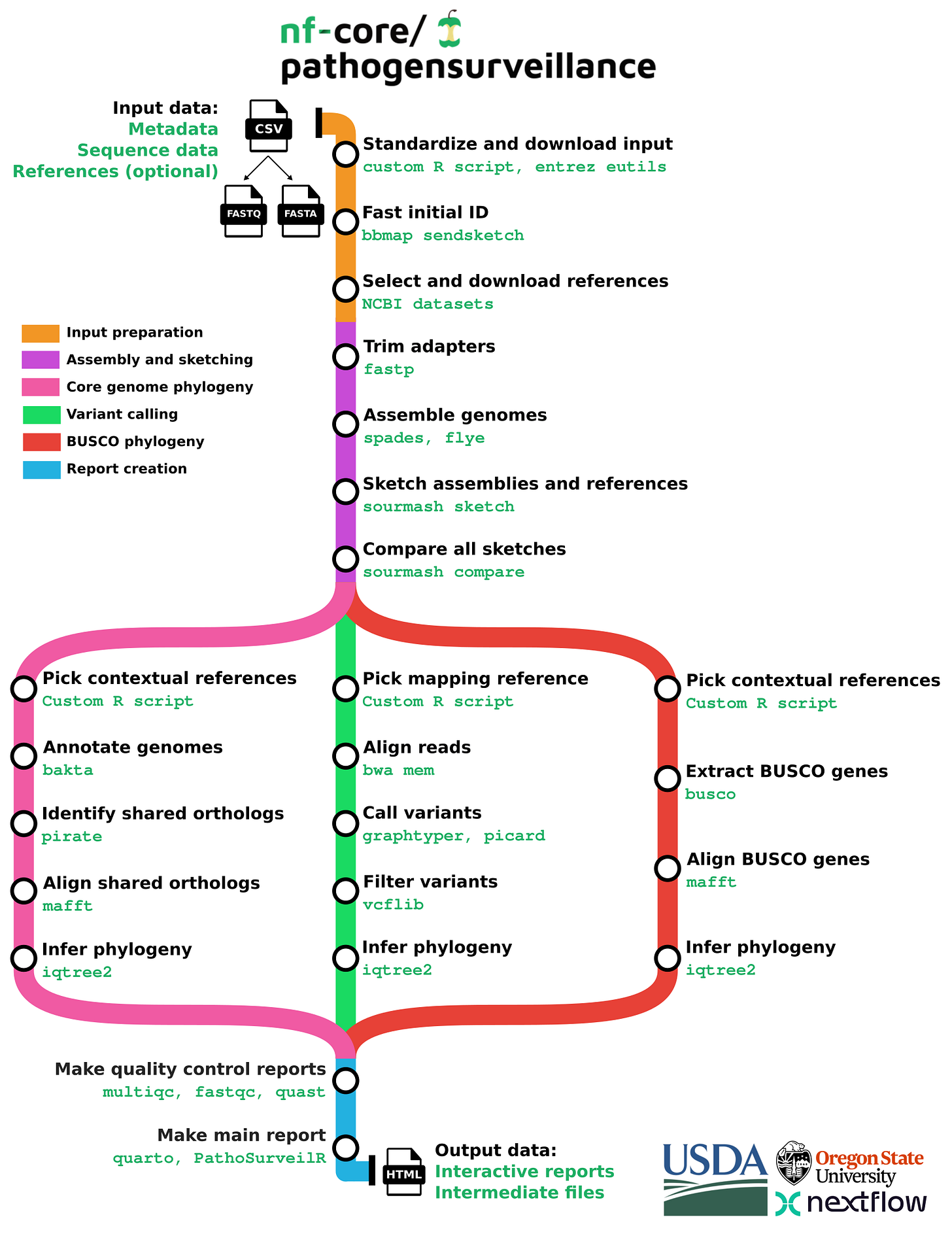
nf-core/pathogensurveillance (paper, code, nf-core pipeline) is a population genomics pipeline for pathogen identification, variant detection, and biosurveillance. The pipeline accepts paths to raw reads for one or more organisms and creates reports in the form of an interactive HTML document.
Finally, a few other papers and preprints that caught my attention this week:
LabOS: The AI-XR Co-Scientist That Sees and Works With Humans
LorBin: efficient binning of long-read metagenomes by multiscale adaptive clustering and evaluation
Benchmarking cell type and gene set annotation by large language models with AnnDictionary
Integrative deep learning of spatial multi-omics with SWITCH
Microbial genomics for antimicrobial resistance ecology and action
Democratizing protein language model training, sharing and collaboration
Generating long deletions across the genome with pooled paired prime editing screens
Privacy-hardened and hallucination-resistant synthetic data generation with logic-solvers
Language models cannot reliably distinguish belief from knowledge and fact
A pre-trained large generative model for translating single-cell transcriptomes to proteomes
Benchmarking of human read removal strategies for viral and microbial metagenomics