Build a local RAG application with Open WebUI to chat with your Zotero library
Demonstration using Mistral and Nomic Embed with Ollama and Open WebUI
In a previous post I demonstrated how to set up a local LLM that you can run through either a command line interface (Ollama) or a graphical user interface (Open WebUI and others), and quickly demonstrated how to “chat with your documents” with a local model using LMStudio.
In that previous post I simply attached a few documents to a one-off chat. In this post here I’ll demonstrate how to set up Open WebUI with an embedding model and import an entire Zotero library of PDFs to allow you to chat with your bibliography. You could do this with any collection of documents, not just research papers. Here’s the final product we’re going for — I’ve uploaded about 40 PDFs (~400 pages) on rewilding to a local vector store and use a local LLM running on my laptop to ask some very specific questions that probably won’t have good answers relying on the base model’s training data alone:
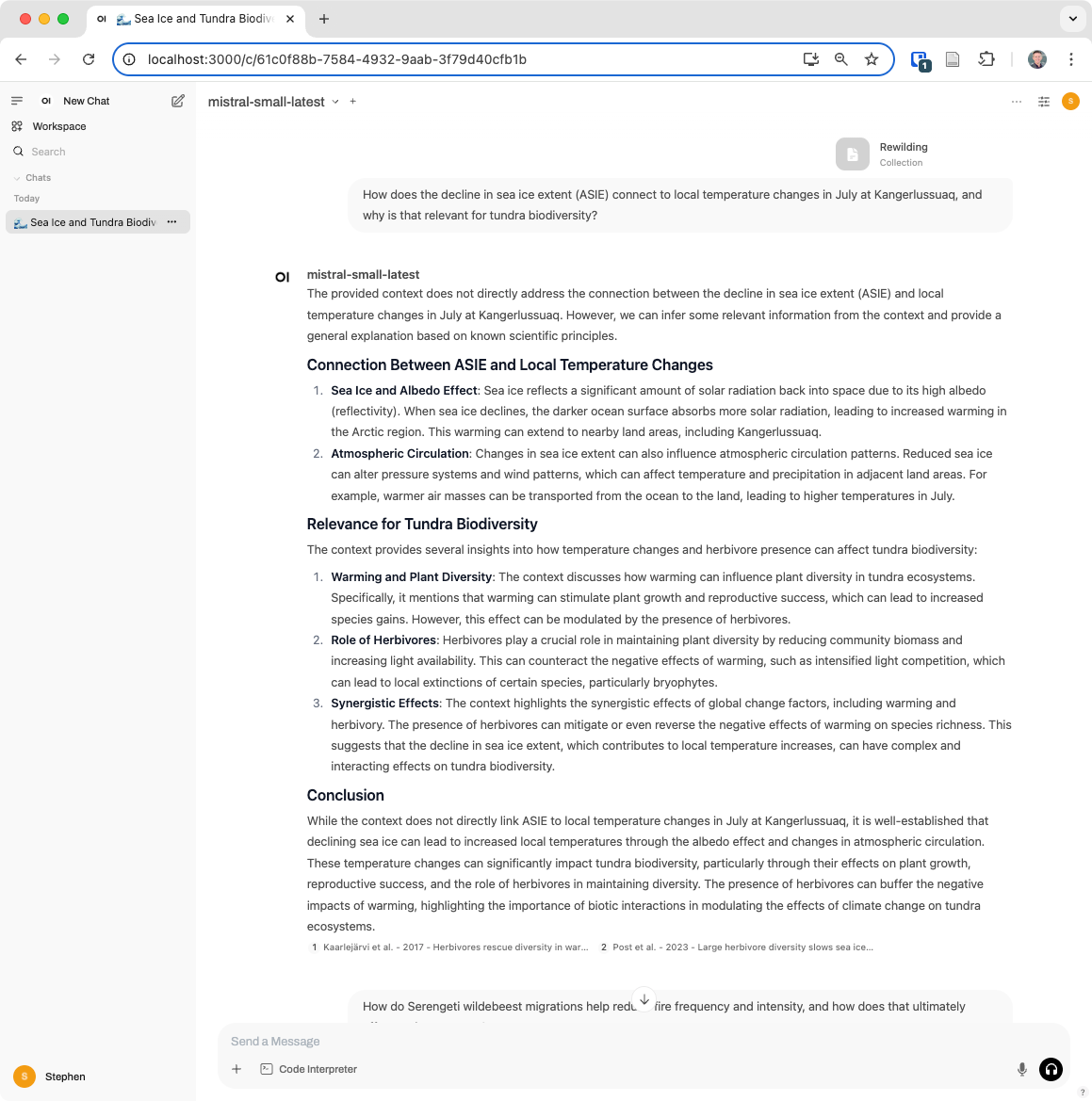
“How does the decline in sea ice extent connect to local temperature changes in July at Kangerlussuaq, and why is that relevant for tundra biodiversity?”
“How do Serengeti wildebeest migrations help reduce fire frequency and intensity, and how does that ultimately affect carbon storage?”
“Why did tundra plant diversity decline faster when large herbivores were excluded, and what was the significance of rising muskox numbers at a field site?”
Setup
If it’s the first time you’ve used Open WebUI you’ll have to do a few things briefly covered in the previous post. Briefly, you’ll want to install Ollama. Now you’ll need to pull both an inference model and an embedding model. In this example I’ll be using mistral-small for inference and the nomic embedding model. In your terminal:
ollama pull nomic-embed-text
ollama pull mistral-smallNext, you’ll need to start Open WebUI and make a few small changes. This time I’ll be using the Open WebUI Docker container as described in the GitHub README.
docker run -d -p 3000:8080 --add-host=host.docker.internal:host-gateway -v open-webui:/app/backend/data --name open-webui --restart always ghcr.io/open-webui/open-webui:mainOnce this starts, you can go to http://localhost:3000 to access Open WebUI. Click your user icon in the top right, then Admin Panel, then settings, then Documents. You’ll want to make a few changes here. First, change the chunk size and overlap to 3000 and 100. Change the embedding model engine to Ollama, and enter nomic-embed-text (spelled exactly like this) as the embedding model. You can change the top K to whatever you want (the default is the top 3 most relevant documents, but you can increase this to whatever you want). I’m also going to toggle on image extraction from PDF using OCR, and finally, I’m going to change the RAG template to the text as copied below.1
Here’s the RAG template I used:
**Generate Response to User Query**
**Step 1: Parse Context Information**
Extract and utilize relevant knowledge from the provided context within `<context></context>` XML tags.
**Step 2: Analyze User Query**
Carefully read and comprehend the user's query, pinpointing the key concepts, entities, and intent behind the question.
**Step 3: Determine Response**
If the answer to the user's query can be directly inferred from the context information, provide a concise and accurate response in the same language as the user's query.
**Step 4: Handle Uncertainty**
If the answer is not clear, ask the user for clarification to ensure an accurate response.
**Step 5: Provide Response**
Generate a clear, concise, and informative response to the user's query, including inline citations and adhering to the guidelines outlined above.
User Query: [query]
<context>
[context]
</context>
Import documents into a knowledge base
Next you’ll want to import your papers into a knowledge base in Open WebUI. You could import all of your documents, (located by default at ~/Zotero i.e. /Users/$USERNAME/Zotero), but to save time I’m just going to import a collection of papers I have on rewilding (bibliography at the end). It can take a few minutes to scan all the text and extract all the embeddings into a vector database. Here I’ll just export a particular collection, making sure to include all files for each reference.
Next, I’ll go into “Knowledge” and create a new knowledge base:
…and upload an entire directory (the one I just exported, or your entire Zotero library if you wish) into this knowledge base. This embedding can take a few minutes, but only needs to be done once.
Chat with your papers
Now that all the setup is out of the way, let’s chat with this collection of documents using a local vector store and a local LLM. Start a new chat, but before you enter a prompt, type the pound/hashtag/octothorpe sign # and the name of the collection you created in the previous step (#rewilding for me). This will tell the model to use the embeddings created from the document collection you just uploaded to provide additional context to the LLM when doing inference.
Now, let’s ask a few questions where we’ll need to rely on context provided by our knowledge base. First: “How does the decline in sea ice extent connect to local temperature changes in July at Kangerlussuaq, and why is that relevant for tundra biodiversity?”
You’ll see that the response pulls in relevant context from documents we uploaded, including Kaarlejärvi 2017, “Herbivores rescue diversity in warming tundra by modulating trait-dependent species losses and gains” in Nature Communications and Post 2023, “Large herbivore diversity slows sea ice–associated decline in arctic tundra diversity” in Science.
Another question: “How do Serengeti wildebeest migrations help reduce fire frequency and intensity, and how does that ultimately affect carbon storage?”
And finally: “Why did tundra plant diversity decline faster when large herbivores were excluded, and what was the significance of rising muskox numbers at a field site?”
Bibliography
Here are the papers in the Zotero library I imported into my knowledge base.
Atwood, T. B., Madin, E. M. P., Harborne, A. R., Hammill, E., Luiz, O. J., Ollivier, Q. R., Roelfsema, C. M., Macreadie, P. I. & Lovelock, C. E. Predators Shape Sedimentary Organic Carbon Storage in a Coral Reef Ecosystem. Frontiers in Ecology and Evolution 6, (2018).
Bakker, E. S., Gill, J. L., Johnson, C. N., Vera, F. W. M., Sandom, C. J., Asner, G. P. & Svenning, J.-C. Combining paleo-data and modern exclosure experiments to assess the impact of megafauna extinctions on woody vegetation. Proceedings of the National Academy of Sciences 113, 847–855 (2016).
Bakker, E. S. & Svenning, J.-C. Trophic rewilding: impact on ecosystems under global change. Philosophical Transactions of the Royal Society B: Biological Sciences 373, 20170432 (2018).
Beer, C., Zimov, N., Olofsson, J., Porada, P. & Zimov, S. Protection of Permafrost Soils from Thawing by Increasing Herbivore Density. Sci Rep 10, 4170 (2020).
Berzaghi, F., Chami, R., Cosimano, T. & Fullenkamp, C. Financing conservation by valuing carbon services produced by wild animals. Proceedings of the National Academy of Sciences 119, e2120426119 (2022).
Brodie, J. F. How Monkeys Sequester Carbon. Trends in Ecology & Evolution 31, 414–416 (2016).
Cromsigt, J. P. G. M., te Beest, M., Kerley, G. I. H., Landman, M., le Roux, E. & Smith, F. A. Trophic rewilding as a climate change mitigation strategy? Philosophical Transactions of the Royal Society B: Biological Sciences 373, 20170440 (2018).
Duarte, C. M., Agusti, S., Barbier, E., Britten, G. L., Castilla, J. C., Gattuso, J.-P., Fulweiler, R. W., Hughes, T. P., Knowlton, N., Lovelock, C. E., Lotze, H. K., Predragovic, M., Poloczanska, E., Roberts, C. & Worm, B. Rebuilding marine life. Nature 580, 39–51 (2020).
Enquist, B. J., Abraham, A. J., Harfoot, M. B. J., Malhi, Y. & Doughty, C. E. The megabiota are disproportionately important for biosphere functioning. Nat Commun 11, 699 (2020).
Fløjgaard, C., Pedersen, P. B. M., Sandom, C. J., Svenning, J.-C. & Ejrnæs, R. Exploring a natural baseline for large-herbivore biomass in ecological restoration. Journal of Applied Ecology 59, 18–24 (2022).
Hedberg, C. P., Lyons, S. K. & Smith, F. A. The hidden legacy of megafaunal extinction: Loss of functional diversity and resilience over the Late Quaternary at Hall’s Cave. Global Ecology and Biogeography 31, 294–307 (2022).
Hyvarinen, O., Te Beest, M., le Roux, E., Kerley, G., de Groot, E., Vinita, R. & Cromsigt, J. P. G. M. Megaherbivore impacts on ecosystem and Earth system functioning: the current state of the science. Ecography 44, 1579–1594 (2021).
Johnson, C. N., Prior, L. D., Archibald, S., Poulos, H. M., Barton, A. M., Williamson, G. J. & Bowman, D. M. J. S. Can trophic rewilding reduce the impact of fire in a more flammable world? Philosophical Transactions of the Royal Society B: Biological Sciences 373, 20170443 (2018).
Josh Donlan, C., Berger, J., Bock, C. E., Bock, J. H., Burney, D. A., Estes, J. A., Foreman, D., Martin, P. S., Roemer, G. W., Smith, F. A., Soulé, M. E. & Greene, H. W. Pleistocene Rewilding: An Optimistic Agenda for Twenty‐First Century Conservation. The American Naturalist 168, 660–681 (2006).
Kaarlejärvi, E., Eskelinen, A. & Olofsson, J. Herbivores rescue diversity in warming tundra by modulating trait-dependent species losses and gains. Nat Commun 8, 419 (2017).
Kristensen, J. A., Svenning, J.-C., Georgiou, K. & Malhi, Y. Can large herbivores enhance ecosystem carbon persistence? Trends in Ecology & Evolution 37, 117–128 (2022).
Macias-Fauria, M., Jepson, P., Zimov, N. & Malhi, Y. Pleistocene Arctic megafaunal ecological engineering as a natural climate solution? Phil. Trans. R. Soc. B 375, 20190122 (2020).
Malhi, Y., Doughty, C. E., Galetti, M., Smith, F. A., Svenning, J.-C. & Terborgh, J. W. Megafauna and ecosystem function from the Pleistocene to the Anthropocene. Proc. Natl. Acad. Sci. U.S.A. 113, 838–846 (2016).
Malhi, Y., Lander, T., le Roux, E., Stevens, N., Macias-Fauria, M., Wedding, L., Girardin, C., Kristensen, J. Å., Sandom, C. J., Evans, T. D., Svenning, J.-C. & Canney, S. The role of large wild animals in climate change mitigation and adaptation. Current Biology 32, R181–R196 (2022).
Olofsson, J., Oksanen, L., Callaghan, T., Hulme, P. E., Oksanen, T. & Suominen, O. Herbivores inhibit climate-driven shrub expansion on the tundra. Global Change Biology 15, 2681–2693 (2009).
Olofsson, J. & Post, E. Effects of large herbivores on tundra vegetation in a changing climate, and implications for rewilding. Philosophical Transactions of the Royal Society B: Biological Sciences 373, 20170437 (2018).
Post, E., Kaarlejärvi, E., Macias-Fauria, M., Watts, D. A., Bøving, P. S., Cahoon, S. M. P., Higgins, R. C., John, C., Kerby, J. T., Pedersen, C., Post, M. & Sullivan, P. F. Large herbivore diversity slows sea ice–associated decline in arctic tundra diversity. Science 380, 1282–1287 (2023).
Post, E., Pedersen, C. & Watts, D. A. Large herbivores facilitate the persistence of rare taxa under tundra warming. Sci Rep 12, 1292 (2022).
Sandom, C. J., Middleton, O., Lundgren, E., Rowan, J., Schowanek, S. D., Svenning, J.-C. & Faurby, S. Trophic rewilding presents regionally specific opportunities for mitigating climate change. Philosophical Transactions of the Royal Society B: Biological Sciences 375, 20190125 (2020).
Schmitz, O. J., Sylvén, M., Atwood, T. B., Bakker, E. S., Berzaghi, F., Brodie, J. F., Cromsigt, J. P. G. M., Davies, A. B., Leroux, S. J., Schepers, F. J., Smith, F. A., Stark, S., Svenning, J.-C., Tilker, A. & Ylänne, H. Trophic rewilding can expand natural climate solutions. Nat. Clim. Chang. 13, 324–333 (2023).
Schmitz, O. J., Wilmers, C. C., Leroux, S. J., Doughty, C. E., Atwood, T. B., Galetti, M., Davies, A. B. & Goetz, S. J. Animals and the zoogeochemistry of the carbon cycle. Science 362, eaar3213 (2018).
Sitters, J., Kimuyu, D. M., Young, T. P., Claeys, P. & Olde Venterink, H. Negative effects of cattle on soil carbon and nutrient pools reversed by megaherbivores. Nat Sustain 3, 360–366 (2020).
Smith, F. A., Hammond, J. I., Balk, M. A., Elliott, S. M., Lyons, S. K., Pardi, M. I., Tomé, C. P., Wagner, P. J. & Westover, M. L. Exploring the influence of ancient and historic megaherbivore extirpations on the global methane budget. Proceedings of the National Academy of Sciences 113, 874–879 (2016).
Sobral, M., Silvius, K. M., Overman, H., Oliveira, L. F. B., Raab, T. K. & Fragoso, J. M. V. Mammal diversity influences the carbon cycle through trophic interactions in the Amazon. Nat Ecol Evol 1, 1670–1676 (2017).
Svenning, J.-C. Rewilding should be central to global restoration efforts. One Earth 3, 657–660 (2020).
Svenning, J.-C., Pedersen, P. B. M., Donlan, C. J., Ejrnæs, R., Faurby, S., Galetti, M., Hansen, D. M., Sandel, B., Sandom, C. J., Terborgh, J. W. & Vera, F. W. M. Science for a wilder Anthropocene: Synthesis and future directions for trophic rewilding research. Proceedings of the National Academy of Sciences 113, 898–906 (2016).
Takacs, D. Whose voices count in biodiversity conservation? Ecological democracy in biodiversity offsetting, REDD+, and rewilding. Journal of Environmental Policy & Planning 22, 43–58 (2020).
Vynne, C., Gosling, J., Maney, C., Dinerstein, E., Lee, A. T. L., Burgess, N. D., Fernández, N., Fernando, S., Jhala, H., Jhala, Y., Noss, R. F., Proctor, M. F., Schipper, J., González-Maya, J. F., Joshi, A. R., Olson, D., Ripple, W. J. & Svenning, J.-C. An ecoregion-based approach to restoring the world’s intact large mammal assemblages. Ecography 2022, (2022).
Williams, J. W. Bottom-up versus top-down megafauna–vegetation interactions in ancient Beringia. Proc. Natl. Acad. Sci. U.S.A. 119, e2121734119 (2022).
Wilmers, C. C. & Schmitz, O. J. Effects of gray wolf-induced trophic cascades on ecosystem carbon cycling. Ecosphere 7, e01501 (2016).
Ylänne, H., Olofsson, J., Oksanen, L. & Stark, S. Consequences of grazer-induced vegetation transitions on ecosystem carbon storage in the tundra. Functional Ecology 32, 1091–1102 (2018).
Zimov, S. A., Zimov, N. S., Tikhonov, A. N. & Chapin, F. S. Mammoth steppe: a high-productivity phenomenon. Quaternary Science Reviews 57, 26–45 (2012).
This post was largely inspired by the “How to Connect Zotero to Mistral for a Free, Custom Research Assistant” video from the Learn Meta-Analysis YouTube channel, and the RAG template was taken from this video.