Weekly Recap (Dec 2024, part 2)
Nextflow to web apps, variant analysis with DRAGEN, synthetic cis-regulatory elements, Nextflow for kinase ID+characterization, language model for single cell analysis, scalable protein design, ...
This week’s recap highlights a new way to turn Nextflow pipelines into web apps, DRAGEN for fast and accurate variant calling, machine-guided design of cell-type-targeting cis-regulatory elements, a Nextflow pipeline for identifying and classifying protein kinases, a new language model for single cell perturbations that integrates knowledge from literature, GeneCards, etc., and a new method for scalable protein design in a relaxed sequence space.
Others that caught my attention include commentary on improving bioinformatics software quality through teamwork, targeted nanopore sequencing for mitochondrial variant analysis, a review on plant conservation in the era of genome engineering, a de novo assembly tool for complex plant organelle genomes, learning to call copy number variants on low coverage ancient genomes, a near telomere-to-telomere phased reference assembly for the male mountain gorilla, a method for optimized germline and somatic variant detection across genome builds, a searchable large-scale web repository for bacterial genomes, and an integer programming framework for pangenome-based genome inference.
Audio generated with NotebookLM. (The hosts were very excited about this issue!)
Deep dive
Cloudgene 3: Transforming Nextflow Pipelines into Powerful Web Services
Paper: Lukas Forer and Sebastian Schönherr. Cloudgene 3: Transforming Nextflow Pipelines into Powerful Web Services. bioRxiv, 2024. DOI: 10.1101/2024.10.27.620456.
I got to meet both Lukas and Sebastian in person at the Nextflow Summit. Lukas gave a talk on nf-test, while Sebastian gave a talk on the Michigan Imputation Server (MIS). MIS is implemented in Nextflow and driven using Cloudgene, and has helped over 12,000 researchers worldwide impute over 100 million samples. This paper describes Cloudgene for turning a Nextflow pipeline into a web service.
TL;DR: Cloudgene 3 provides a user-friendly platform to convert Nextflow pipelines into scalable web services, allowing scientists to deploy and run complex bioinformatics workflows without requiring web development expertise.
Summary: Cloudgene 3 addresses the challenge of deploying Nextflow pipelines as scalable web services, allowing researchers to leverage computational workflows without the need for technical setup or coding. The platform simplifies the transformation of Nextflow pipelines into “Cloudgene apps,” which include user-friendly interfaces and allow for seamless dataset management, job monitoring, and data security. By supporting features like workflow chaining and dataset integration, Cloudgene 3 enables collaborative and flexible use of pipelines across various scientific domains, from genomics to proteomics. This tool expands accessibility to complex analyses, facilitating data sharing and enhancing reproducibility, and has already been implemented in large-scale services like the Michigan Imputation Server. Its open accessibility and adaptable deployment model (cloud or local infrastructure) highlight its utility for bioinformatics workflows.
Methodological highlights:
Converts Nextflow pipelines into web services with a few simple steps, creating portable “apps” that include metadata, input/output parameters, and multi-step workflows.
Integrates real-time status updates and error handling for Nextflow tasks, leveraging a unique secret URL for each task to monitor progress.
Supports cloud platforms and local installations, providing compatibility with engines like Slurm and AWS Batch and storage options like AWS S3.
New tools, data, and resources:
Cloudgene 3 platform: Free platform available at cloudgene.io.
Cloudgene 3 source code: https://github.com/genepi/cloudgene3.


Comprehensive genome analysis and variant detection at scale using DRAGEN
Paper: Behera, S., et al. Comprehensive genome analysis and variant detection at scale using DRAGEN. Nature Biotechnology, 2024. DOI: 10.1038/s41587-024-02382-1.
DRAGEN was a godsend in a previous job. I needed a turnkey variant calling solution that was fast. I bought an on-prem DRAGEN FPGA server, which was capable of taking you from FASTQ files to VCF in ~30 minutes for a 30X human whole genome. Illumina has previously published white papers on DRAGEN’s speed and accuracy. The publication in Nature Biotechnology engendered some interesting discussion online. On one hand, the paper was a pleasure to read, and the benchmarks are compelling and well done. On the other, the method isn’t available to explore, reproduce, understand in detail, or build upon. Which raises the question — should this have been a peer-reviewed publication in the scientific record? Or should this just have been another white paper? At some point “papers” hawking some new and improved closed source method are thinly veiled advertisements stamped with the approval of peer review. I think there should be some place in the scientific literature for papers like this describing a closed-source method, but where benchmarks are independently evaluated by a team of peer reviewers. I just don’t know what that looks like in the current landscape of peer reviewed papers versus a vendor’s white paper.
TL;DR: DRAGEN is a high-speed, highly accurate genomic analysis platform for variant detection, leveraging hardware acceleration, pangenome references, and machine learning. It outperforms traditional tools across variant types (SNVs, indels, SVs, CNVs, STRs) and is designed for large-scale, clinical genomics applications.
Summary: This study presents DRAGEN, a platform that uses accelerated hardware and sophisticated algorithms to enable comprehensive variant detection at unprecedented speed and accuracy. By integrating pangenome references and optimizing for all major variant classes, DRAGEN achieves high concordance in identifying complex and diverse genomic variants, even in challenging regions. Benchmarking across 3,202 genomes from the 1000 Genomes Project highlights DRAGEN’s scalability and its advantages over traditional methods like GATK and DeepVariant, especially for clinically relevant genes. The platform’s robust performance across SNVs, SVs, CNVs, and STRs allows for large-cohort analyses critical for population-scale genomics and clinical diagnostics, facilitating variant discovery in diseases with both common and rare genetic underpinnings.
Methodological highlights:
Uses pangenome references to enhance alignment accuracy and variant detection across diverse populations.
Optimized for rapid, parallel processing of SNVs, indels, CNVs, and STRs with an average processing time of ~30 minutes per genome.
Employs machine learning-based filtering to reduce false positives and improve accuracy in variant calling.
Integration of ExpansionHunter for STR analysis and specialized callers for pharmacogenomic variants (e.g., CYP2D6, SMN) ensures reliable detection in medically significant genes.

Machine-guided design of cell-type-targeting cis-regulatory elements
Paper: Gosai, S. J., et al. Machine-guided design of cell-type-targeting cis-regulatory elements. Nature, 2024. DOI: 10.1038/s41586-024-08070-z.
TL;DR: This paper introduces a platform for designing synthetic cis-regulatory elements (CREs) with programmed cell-type specificity using a deep-learning-based model called Malinois, combined with a computational design tool, CODA, and massively parallel reporter assays (MPRAs) for validation.
Summary: This study presents a framework for designing synthetic CREs that drive gene expression specifically in desired cell types. Using Malinois, a deep convolutional neural network trained on MPRA data from human cells, the researchers predict CRE activity and design synthetic elements targeting specific cell lines. The CODA (Computational Optimization of DNA Activity) platform then iteratively refines these designs to achieve high specificity, which is validated in vitro across multiple cell types and in vivo in mice and zebrafish. By outperforming natural CREs in specificity and robustness, these synthetic elements could significantly enhance targeted gene therapy approaches, especially by providing tools for precise gene expression control in therapeutic and research applications. The framework expands our capacity to engineer regulatory DNA for complex tissue-specific requirements, advancing possibilities for both biomedical research and gene therapy.
Methodological highlights:
Malinois CNN model predicts cell-type-specific CRE activity directly from DNA sequences, validated with MPRA-based data in K562, HepG2, and SK-N-SH cells.
CODA optimization platform iteratively adjusts CRE sequences to increase cell-type specificity, employing algorithms such as Fast SeqProp for efficient sequence design.
High-throughput MPRA validates the activity of 77,157 synthetic and natural CRE sequences across cell types, showing superior specificity in synthetic CREs.
New tools, data, and resources:
Code availability: https://github.com/sjgosai/boda2 (yes, this is the CODA repo, which is named “boda2” for “legacy reasons”).
Data availability: All the data used in the study is described in the data availability section of the paper.

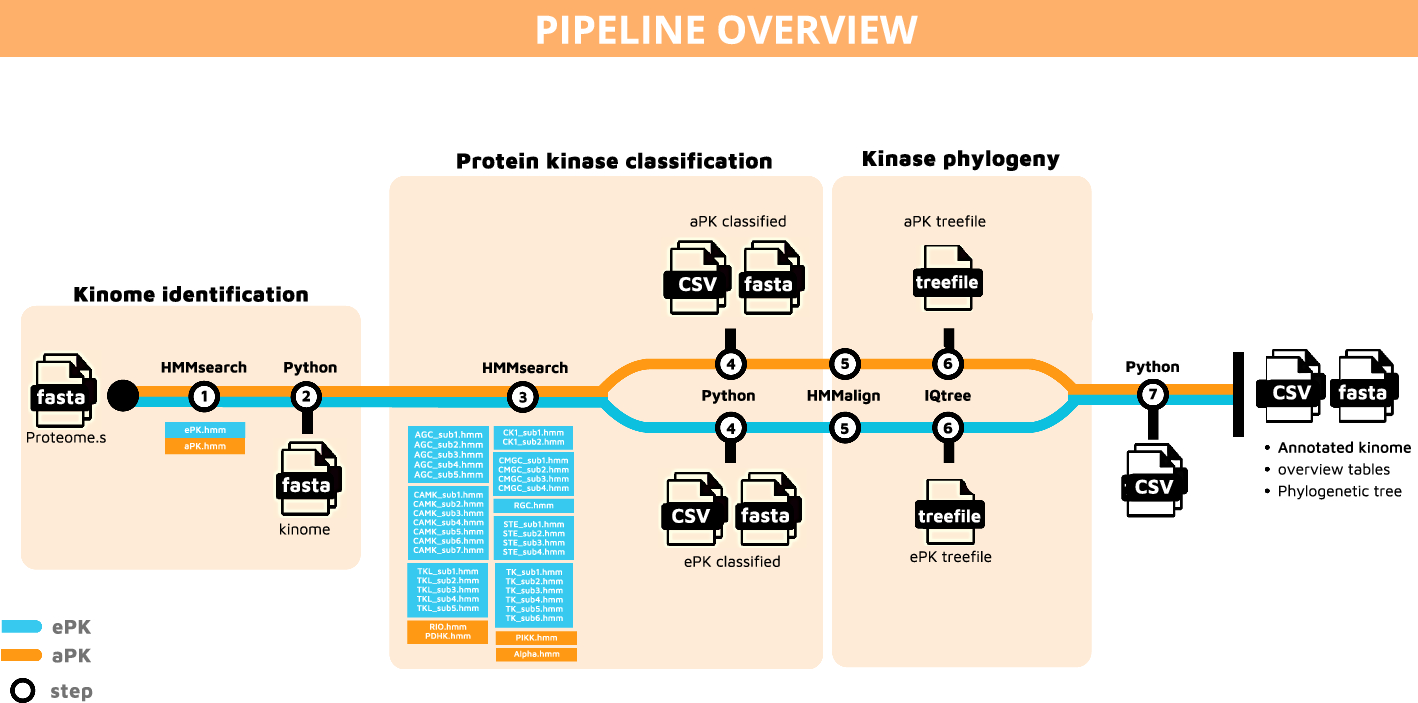
KiNext: a portable and scalable workflow for the identification and classification of protein kinases
Paper: Hellec, E., et al. KiNext: A Portable and Scalable Workflow for the Identification and Classification of Protein Kinases. BMC Bioinformatics, 2024. DOI: 10.1186/s12859-024-05953-w.
TL;DR: KiNext is a Nextflow-based pipeline for identifying and classifying protein kinases (kinome) from annotated genomes, enabling reproducible analysis and classification of kinase families across species.
Summary: Protein kinases are crucial for cellular signaling and adaptation, and identifying the full kinome of an organism can reveal insights into its physiological and adaptive capabilities. KiNext automates this process, applying Hidden Markov Models (HMMs) to detect both eukaryotic and atypical protein kinases from genomic data and classifying them into known kinase families. Validated against two model species, Crassostrea gigas and Ostreococcus tauri, KiNext identified previously unclassified kinases and achieved enhanced classification accuracy compared to earlier methods. The tool is particularly valuable for large-scale genome projects, such as the Earth BioGenome Project, as it ensures reproducible kinome analysis in alignment with FAIR data principles. By allowing user-provided HMMs, it can be adapted for diverse taxa, making it versatile for comparative kinome studies.
Methodological highlights:
Uses Nextflow and Singularity containers for reproducible, scalable analysis of kinomes across computing environments.
Combines HMM models for specific kinase groups with automatic phylogenetic analysis of ePKs and aPKs using IQTree, facilitating precise classification.
Outputs include detailed summary tables of identified kinases, with options for further validation using structural prediction tools like AlphaFold and Foldseek.
New tools, data, and resources:
Code availability: On GitLab at https://gitlab.ifremer.fr/bioinfo/workflows/kinext. Note that this is licensed under the AGPL, not the regular GPL. Meaning that the source code availability trigger in the GPL also applies if the pipeline is offered over a network as a service.
scGenePT: Is language all you need for modeling single-cell perturbations?
Paper: Istrate, A.-M., et al. scGenePT: Is language all you need for modeling single-cell perturbations? bioRxiv, 2024. https://doi.org/10.1101/2024.10.23.619972.
This work from researchers at the Chan Zuckerberg Initiative was also covered a recent issue of the Bits In Bio Weekly newsletter (which, by the way, if you haven’t subscribed to the newsletter or joined the BiB Slack, you should). Of note the text embeddings use GPT 3.5, which is already outdated. Unfortunately the code and model checkpoints aren’t available, and “will be made available upon publication.”
TL;DR: This study introduces scGenePT, an enhanced single-cell gene perturbation model combining biological and language-based representations to predict gene expression outcomes. By incorporating gene knowledge from scientific literature, it improves over traditional models like scGPT in complex perturbation settings.
Summary: In advancing single-cell biology, predicting the effects of gene perturbations is essential. Traditional models rely on experimental data, such as single-cell RNA sequencing counts, but scGenePT integrates language embeddings from scientific resources (e.g., NCBI, UniProt, Gene Ontology) to enrich gene representations. The findings suggest that adding textual gene representations enhances the predictive power for single- and two-gene perturbations, especially where gene interactions yield non-additive effects. This approach is crucial for handling diverse perturbation scenarios, with significant implications for precision medicine, as it facilitates a more nuanced understanding of gene interactions under various conditions. The study validates the integration approach with superior performance over models relying solely on experimental data, demonstrating that language representations can serve as a valuable prior in biological modeling.
Methodological highlights:
scGenePT extends scGPT by adding gene-specific language embeddings sourced from scientific literature, providing both additive and complementary information for perturbation modeling.
Uses GPT-3.5 embeddings to align knowledge sources like NCBI gene summaries, UniProt protein data, and Gene Ontology annotations.
Employs a Transformer-based architecture where language and experimental data embeddings converge at the gene representation level, followed by a Transformer encoder-decoder setup for predictive modeling.
Code availability: Unfortunately the code and model checkpoints aren’t available, and “will be made available upon publication.”

Scalable protein design using optimization in a relaxed sequence space
Paper: Frank, C., et al. Scalable Protein Design Using Optimization in a Relaxed Sequence Space. Science, 2024. DOI: 10.1126/science.adq1741.
Protein design is important in medicine and biotechnology because it enables the creation of synthetic proteins tailored for specific functions, such as therapeutic agents that can target diseases more effectively or robust enzymes for diagnostics. In the area I work in (de-extinction), designed proteins can help reconstitute essential biological functions in revived or closely related species, potentially aiding in biodiversity restoration and ecological balance.
TL;DR: This paper presents a new pipeline for protein design using relaxed sequence optimization (RSO), which enables efficient gradient-based design of large and complex protein structures. This approach yields high-quality protein backbones and supports applications in synthetic protein-protein interactions.
Summary: The study introduces Relaxed Sequence Optimization (RSO), a gradient-descent method applied in a “relaxed” sequence space that enables rapid convergence in protein design without the constraints of one-hot encoding. By iteratively optimizing loss functions within AlphaFold2’s predictive space, RSO creates diverse protein backbones, later refined by ProteinMPNN to ensure foldability into intended structures. This approach was experimentally validated with over 100 designed proteins, including large monomers and heterodimers, highlighting its scalability and structural accuracy for de novo proteins up to 1000 amino acids. The RSO pipeline, which allows for custom loss functions tailored to specific design goals, represents a significant advance in both design speed and application scope, facilitating the development of stable, large-scale proteins relevant to fields like structural biology and synthetic biochemistry.
Methodological highlights:
RSO operates in a relaxed sequence space, optimizing backbones without forcing discrete amino acid representations, allowing for smooth gradient transitions.
Integrates ProteinMPNN for generating foldable protein sequences that match RSO-designed backbones, achieving stable high-resolution structures in experimental validation.
Customizable loss functions support specific design tasks, such as helical content reduction and binder-targeting configurations, without retraining.
New tools, data, and resources:
Code availability: Available on GitHub: github.com/sokrypton/ColabDesign
Data availability: Experimental data, including TEM images and crystal structures, are archived on Figshare at doi.org/10.6084/m9.figshare.27009724, with additional cryo-EM and crystal models in PDB and EMDB.

Other papers of note
Improving bioinformatics software quality through teamwork https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btae632/7831429
Targeted nanopore sequencing using the Flongle device to identify mitochondrial DNA variants https://www.nature.com/articles/s41598-024-75749-8
Review: Plant conservation in the age of genome editing: opportunities and challenges https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03399-0
Oatk: a de novo assembly tool for complex plant organelle genomes https://www.biorxiv.org/content/10.1101/2024.10.23.619857v1
LYCEUM: Learning to call copy number variants on low coverage ancient genomes https://www.biorxiv.org/content/10.1101/2024.10.28.620589v2
A near telomere-to-telomere phased reference assembly for the male mountain gorilla https://www.biorxiv.org/content/10.1101/2024.10.28.620258v1
StableLift: Optimized Germline and Somatic Variant Detection Across Genome Builds https://www.biorxiv.org/content/10.1101/2024.10.31.621401v1
BakRep – a searchable large-scale web repository for bacterial genomes, characterizations and metadata https://doi.org/10.1099/mgen.0.001305
Integer programming framework for pangenome-based genome inference https://www.biorxiv.org/content/10.1101/2024.10.27.620212v1.full
