Weekly recap (Sep 26, 2025)
Apple gets into protein folding, parsing R/Markdown and Quarto, vibe-coding an R package, AI in biosecurity and writing, nothing about AI is inevitable, conservation applications of de-extinction tech
Happy Friday, colleagues. September has gone by at warp speed. It’s the end of the week and once again I’m going through my long list of idle browser tabs trying to catch up where I can.
Apple is entering the protein folding arena with a new preprint on arXiv this week: SimpleFold: Folding Proteins is Simpler than You Think. The code is open-source, MIT licensed, on GitHub (apple/ml-simplefold) and there’s a demo ipynb you can play with. From the README:
We introduce SimpleFold, the first flow-matching based protein folding model that solely uses general purpose transformer layers. SimpleFold does not rely on expensive modules like triangle attention or pair representation biases, and is trained via a generative flow-matching objective. We scale SimpleFold to 3B parameters and train it on more than 8.6M distilled protein structures together with experimental PDB data. To the best of our knowledge, SimpleFold is the largest scale folding model ever developed. On standard folding benchmarks, SimpleFold-3B model achieves competitive performance compared to state-of-the-art baselines
R Weekly 2025-W39: Parse RMarkdown, Vibe Code, Quality Control, and this week’s Data Science with R newsletter.
Maëlle Salmon, Christophe Dervieux, and Zhian N. Kamvar on the rOpenSci blog: All the Ways to Programmatically Edit or Parse R Markdown / Quarto Documents (tinkr, md4r, pandoc, parseqmd, parsermd, lightparser, xmlparsedata, treesitter).
This article from Jonathan Carroll was highlighted on the R Weekly newsletter above: I Vibe Coded an R Package, and it ... actually works?!?
Speaking of vibe coding… An interesting market is emerging: Vibe Coding Cleanup as a Service. Claude Code, Positron Assistant, OpenAI Codex, Cursor, Windsurf, and any number of other agentic AI coding tools can easily let you vibe code yourself into an abyss. The cleanup economy is real. People are building careers around fixing vibe coded slop, and demand is so high that VibeCodeFixers.com launched as a dedicated marketplace.
Related, an essay from Can Elma: AI Was Supposed to Help Juniors Shine. Why Does It Mostly Make Seniors Stronger? This essay takes a look at where AI tools shine versus where they fall short, arguing that AI’s strengths helps senior developers much more than juniors.
And a few more recent reports on companies not seeing any meaningful return on investment in embracing AI in the workplace.
404 Media: AI ‘Workslop’ Is Killing Productivity and Making Workers Miserable
Harvard Business Review: AI-Generated “Workslop” Is Destroying Productivity
Financial Times: America’s top companies keep talking about AI — but can’t explain the upsides
A new preprint: A biosecurity agent for life cycle LLM biosecurity alignment.
Related, over the last few weeks I’ve been quietly posting articles in the AI x Bio space, specifically around biosecurity. Richard Moulange, AI-Biosecurity Policy Manager at the The Centre for Long-Term Resilience posted his first essay on his new newsletter, AI agents will upset plans to safeguard narrow AI–bio tools. AI-bio tools are transforming science but carry serious dual-use risks, prompting safeguards such as harm refusal, user verification, and managed access. The central concern raised in this article is that rapidly advancing AI agents may soon bypass these protections by reproducing safeguarded models from published methods, making powerful capabilities freely available to anyone. This essay reminds me of the central plot device in AI 2027 exploring a near-term future of rapidly advancing AI agents.
Decoding Bio: BioByte 132 weekly highlights. Decoding Bio is a “community of writers, scientists, founders, and funders focused on exploring the boundaries of bio, and elevating an understanding of cutting edge developments in what the kids are calling biotech.” I love their weekly recaps. This week’s issue covers a few super interesting new preprints and papers (below).
Generative genomics accurately predicts future experimental results [Koytiger et al., bioRxiv, September 2025]
Why it matters: RNAseq profiles of different experimental types are laborious and expensive to acquire. Koytiger and Walsh et al. develop GEM-1, which leverages a deep latent-variable model trained on mined RNAseq data with LLM-cleaned metadata to enable scientists to generate specialized transcriptomic datasets in silico.
Engineered prime editors with minimal genomic errors [Chauhan et al., Nature, September 2025]
Why it matters: Prime editors have emerged as a supremely powerful class of genome editors, capable of carrying out insertions, deletions, and substitution edits in DNA without requiring double-stranded breaks. However, the technology is still prone to insertion and deletion errors during the editing process which potentially undermine its overall effectiveness. In this paper, the authors focus on the competition between the 3’ end of the nicked strand compared to the tightly bound 5’ end as a reason for indel errors and propose mutating the Cas9-DNA interface to promote binding of the edited strand. This rational design approach demonstrates a significant reduction in indel errors and paves the way to improve the accuracy of other prime editors.
Fast Company interviews UVA SDS faculty member and historian Mar Hicks on why nothing about AI is inevitable.
Epoch AI report: What will AI look like in 2030?
Science: Can ChatGPT help science writers? Answer: 🤷♂️
The results were mixed. The LLM did summarize scientific findings in language that was accessible to nonexperts (it avoided technical terms and jargon, for example). It also effectively summed up commentary content, such as Science’s Policy Forums, for a lay audience. However, it tended to sacrifice accuracy for simplicity. The ChatGPT Plus summaries required rigorous fact-checking by SciPak writers. Also, extensive editing for hyperbole was needed. For example, ChatGPT Plus had a fondness for using the word “groundbreaking.” It also struggled to highlight more than one result from multifaceted studies. When asked to summarize two papers at once, it could only cover the first of the two submitted. It ultimately defaulted to jargon if challenged with research particularly dense in information, detail, and complexity.
Surya Ganguli’s reading list from an explainable AI + neuroscience course at Stanford. Not my specialty, but from an outsider’s perspective it looks fairly comprehensive.
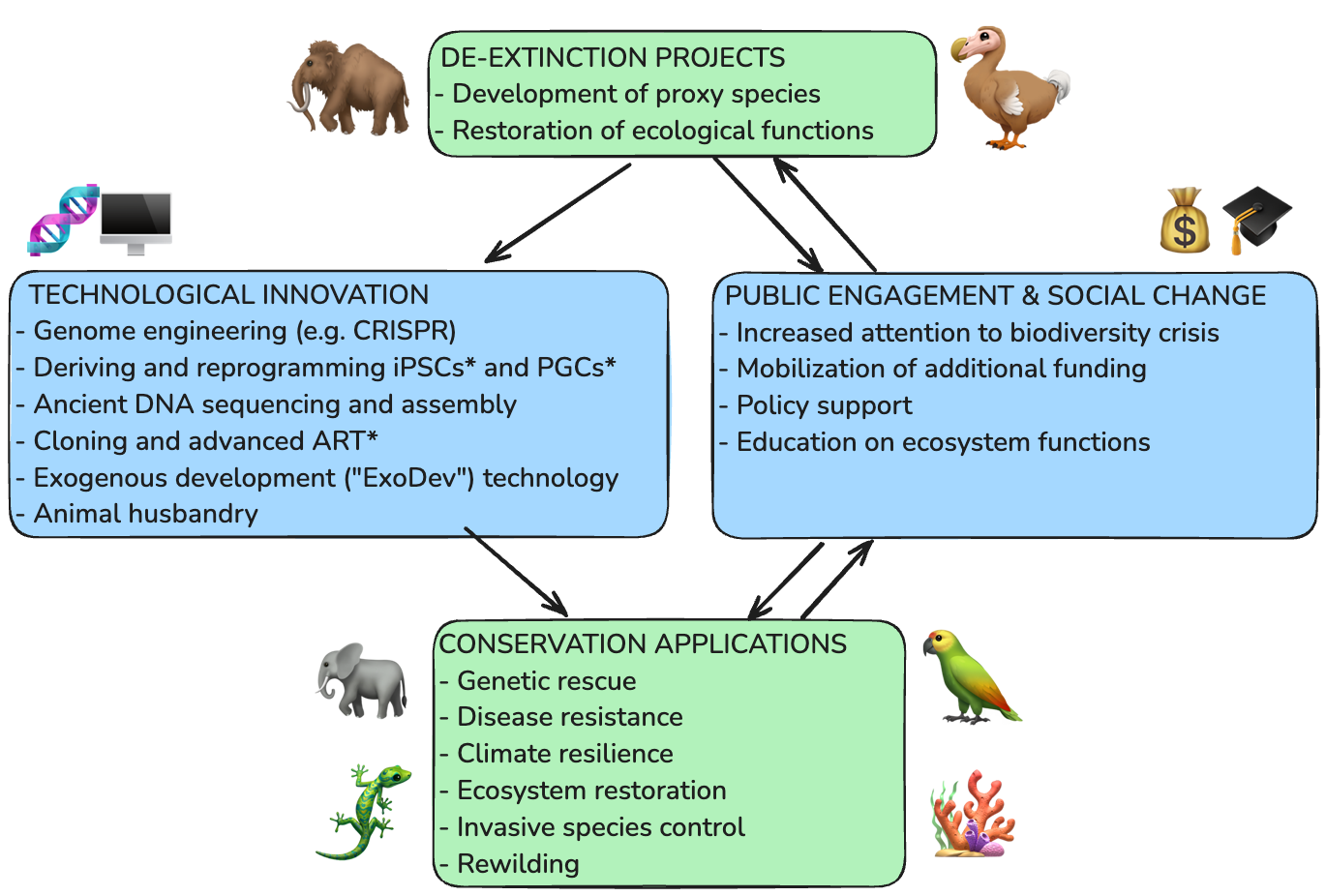
Some shameless self-promotion here. This week I published a review paper in Journal of Heredity that I started while at Colossal: De-extinction technology and its application to conservation. Co-authors include Colossal’s chief biology officer Andrew Pask, avian species director Anna Keyte, and chief science officer Beth Shapiro. The paper reviews many of the conservation applications of technology being further developed for de-extinction research.
Finally, a few other papers and preprints that caught my attention this week:
Opportunities and considerations for using artificial intelligence in bioinformatics education
PatchWorkPlot: simultaneous visualization of local alignments across multiple sequences
Genomics will forever reshape forensic science and criminal justice
Reference genome choice compromises population genetic analyses
De novo Design of All-atom Biomolecular Interactions with RFdiffusion3
The Illusion of Readiness: Stress Testing Large Frontier Models on Multimodal Medical Benchmarks