Weekly Recap (Nov 14, 2025)
posit::conf(2025) & Nextflow Summit talks, bioRxiv+medRxiv AI reviews, R updates (R Data Scientist, R weekly), Claude Scientific Skills, Python package development for R users, new papers & preprints
All the talks from posit::conf(2025) and the 2025 Nextflow Summit are on Youtube. I embedded all the recordings into a single page for each:
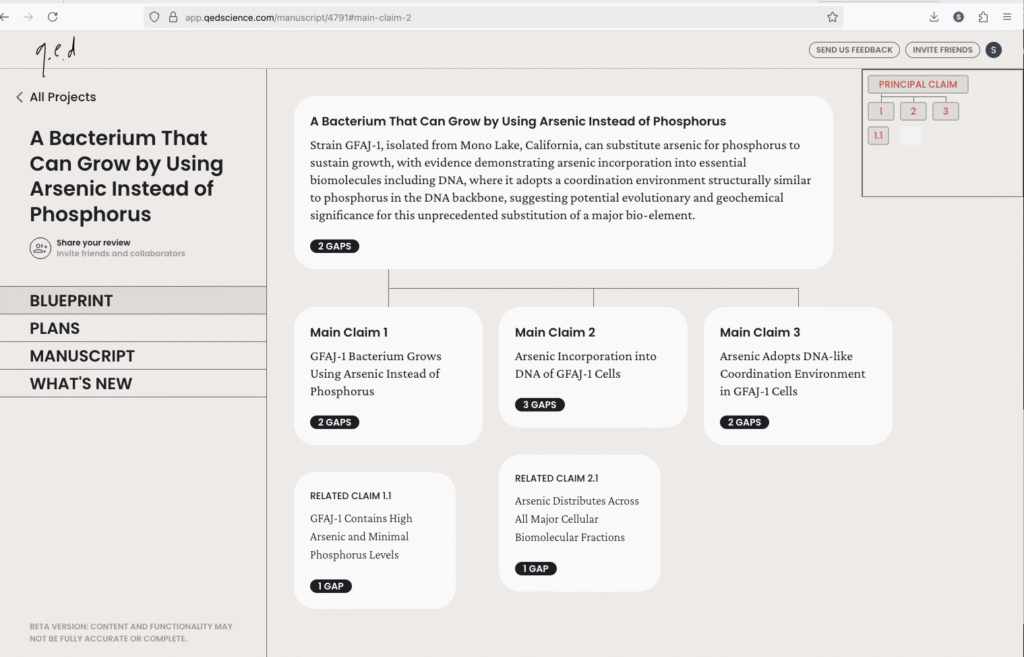
openRxiv (bioRxiv+medRxiv) launches a partnership with q.e.d. to enable automated AI reviews of preprints.
R Weekly 2025-W46: Writing your own ggplot2 geoms, reverse dependency check speedrun: a data.table case study, NYC election.
R Data Scientist 2025-11-11: Community & news, interactive visuals, geospatial analysis, statistical methods, ML & modeling, academic research.
The AI engineer 2025-11-12: Company engineering blogs, open models & architectures, agents & RL, retrieval & RAC, systems & deployment, theory & foundations, academic research.
Yohann Mansiaux: Python Package Development for R developers, Part 1 & Part 2.
The Principles of Diffusion Models: A 470-page monograph from OpenAI, Sony AI, and Stanford University.
pkgdown 2.2.0 released with some new features to make your docs LLM friendly:
This version of pkgdown has one major change: a new
pkgdown::build_llm_docs()function that automatically creates files that make it easier for LLMs to read your documentation. Concretely, this means two things:
You’ll get an
llms.txtat the root directory of your site.llms.txtis an emerging standard that provides an easy way for an LLM to get an overview of your site. pkgdown creates an overview by combining your README, your function index, and your article index: this should give the LLM a broad overview of what your package does, along with links to find out more.Every existing
.htmlon your site gets a corresponding.mdfile. These are generally easier for LLMs to understand because they contain just the content of the site, without any extraneous styling.
Sara Altman & Simon Couch: Posit 2025-11-07 AI Newsletter: Introspection in LLMs, Positron Assistant support for custom model providers, side::kick() agent for RStudio, mcptools, R+AI conference, emergent capabilities.
Ulkar Aghayeva and Asimov Press: What Makes an Experiment Beautiful? A beautiful experiment is not just a reflection of human ingenuity but also efficient science.
K-visa: China’s version of the H-1B aims to attract foreign tech workers.
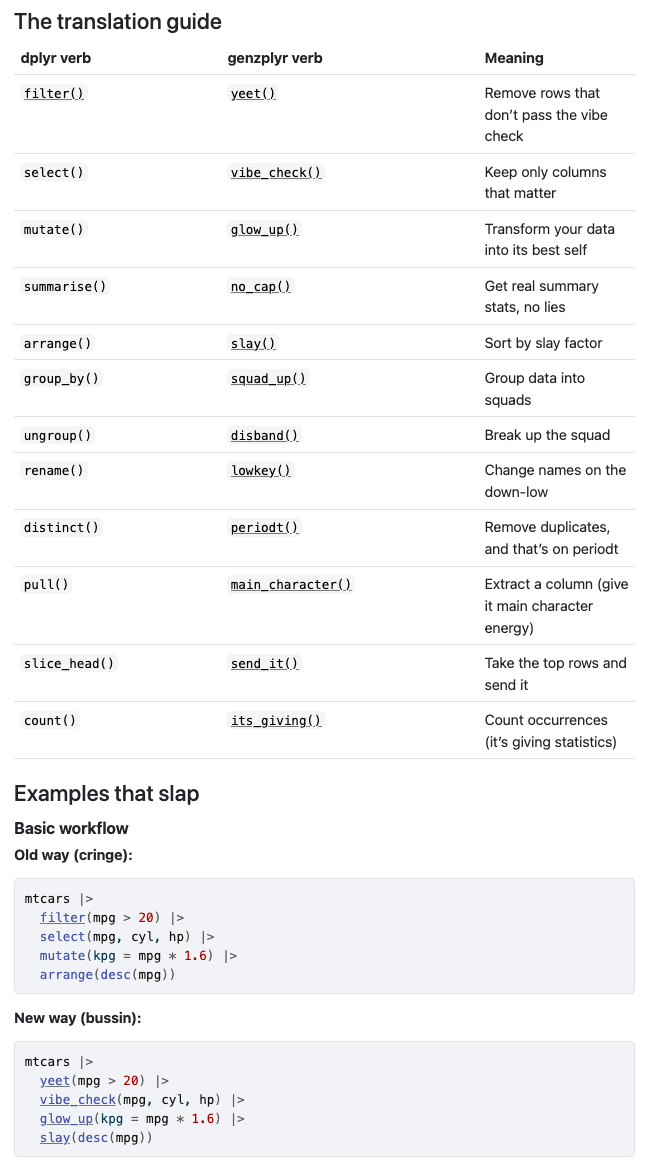
And, because it’s Friday — genzplyr: dplyr but make it bussin fr fr no cap.
Finally, a few other papers and preprints that caught my attention this week:
Decoding Cell Fate: Integrated Experimental and Computational Analysis at the Single-Cell Level
BioWorkflow: Retrieving comprehensive bioinformatics workflows from publications
Site-specific DNA insertion into the human genome with engineered recombinases
Multi-agent AI enables evidence-based cell annotation in single-cell transcriptomics
Nextpie: a web-based reporting tool and database for reproducible nextflow pipelines
Multimodal learning enables chat-based exploration of single-cell data
Estimation and mapping of the missing heritability of human phenotypes