Weekly Recap (March 2025, part 2)
AI agents for biomedical reasoning, species tree inference, a spatial transcriptomics foundation model, biomedical research with LLMs, mapping cells through time and space, epigenetic clocks, SVs, ...
This week’s recap highlights ESCARGOT, an AI agent for biomedical knowledge graphs and reasoning, CASTER for direct species tree inference from whole-genome alignments, the scGPT-spatial foundation model for spatial transcriptomics, the BioChatter platform for biomedical research applications with LLMs, moscot for mapping cells through time and space, and two reviews: one on epigenetic clocks and another on structural variation in the human genome.
Others that caught my attention include a chemical reprogramming system to generate human pluripotent stem cells, massively parallel characterization of transcriptional regulatory elements, a universal droplet microfluidics-based combinatorial indexing strategy for massive-scale multimodal single-cell sequencing, what LLMs know versus what people think they know, challenges in detecting ecological interactions using sedimentary ancient DNA data, overcomming challenges associated with broad sharing of human genomic data, organization of cis-regulatory elements, global patterns in the human gut microbiome, a genome-wide atlas of human cell morphology, and a sobering meta-analysis showing that immediate conservation action is needed to halt genetic diversity loss.
Deep dive
ESCARGOT: an AI agent leveraging large language models, dynamic graph of thoughts, and biomedical knowledge graphs for enhanced reasoning
Paper: Matsumoto, et al. "ESCARGOT: An AI Agent Leveraging Large Language Models, Dynamic Graph of Thoughts, and Biomedical Knowledge Graphs for Enhanced Reasoning," Bioinformatics, 2025. DOI: 10.1093/bioinformatics/btaf031.
This paper comes out of Jason Moore’s lab at the Center for Artificial Intelligence Research and Education, Cedars Sinai Medical Center. Jason was my PhD mentor’s PhD mentor, my “academic grandfather.” I was excited to read this, as I’ve been thinking about building a RAG app for a genomics research application. This paper was published Jan 22, 2025, before the widespread availability of new reasoning models like DeepSeek-R1 and OpenAI’s o3-mini models. I’m interested to see how these results stand up if benchmarked against some of these newer reasoning / chain-of-thought models.
TL;DR: This paper introduces ESCARGOT, an AI agent that fuses LLMs with a dynamic Graph of Thoughts (GoT) and biomedical knowledge graphs to reduce hallucinations and improve precision in biomedical reasoning tasks. It outperforms standard RAG in complex multi-hop biomedical queries.
Summary: ESCARGOT is a new Python-based AI agent that enhances LLM-driven biomedical problem-solving by integrating dynamic reasoning with Graph of Thoughts (GoT) and structured querying of knowledge graphs. It dynamically generates executable reasoning strategies using Cypher queries to databases like AlzKB, allowing fine-grained multi-step inference. The GoT structure allows modular execution, self-debugging, and Python-based computation between knowledge retrieval steps. Benchmarking with GPT-4o Mini shows ESCARGOT drastically outperforms standard RAG approaches in multi-hop reasoning tasks (e.g., 88.4% accuracy in 1-hop open-ended biomedical queries vs. 55.5% with RAG and 4.2% for GPT-4o mini), highlighting its potential to support complex biomedical knowledge extraction and decision-making.
Methodological Highlights:
Dynamic Graph of Thoughts (GoT): Strategy generation + Python-executable graph for multi-step reasoning; adaptive and resilient to query failures.
Hybrid Knowledge Retrieval: Prioritizes Cypher queries to structured knowledge graphs (e.g., Neo4j, Memgraph), with vector search fallback for unstructured data.
Self-Debugging Python Execution: Executes Python code blocks as nodes in GoT; detects errors, rewrites code, and retries autonomously.
New Tools, Data, and Resources:
Code: Available at https://github.com/EpistasisLab/ESCARGOT. Written in Python, MIT license.
Data (AlzKB Knowledge Graph): Alzheimer’s research-oriented graph database (Memgraph); used for benchmarking. Access at https://alzkb.ai/.


CASTER: Direct species tree inference from whole-genome alignments
Paper: Zhang, et al. "CASTER: Direct species tree inference from whole-genome alignments," Science, 2025. https://doi.org/10.1126/science.adk9688.
From Zoonomia to the Earth BioGenome Project whose ambitious goal is to “sequence, catalog and characterize the genomes of all of Earth’s eukaryotic biodiversity,” many new genomes are being sequenced providing ample data for species-level phylogenetic reconstruction.
TL;DR: This paper introduces CASTER, a new site-based phylogenomic method that infers species trees directly from whole-genome alignments without needing pre-defined gene trees or recombination-free loci. It’s more accurate, faster, and scales better than existing methods, handling hundreds of whole genomes while also revealing local discordance patterns across the genome.
Summary: CASTER is a novel species tree inference method that leverages site patterns in whole-genome alignments, avoiding the need to define individual gene trees or recombination-free loci. It is built on coalescence theory and introduces a scoring system for quartets of species based on site patterns, enabling efficient and accurate inference of species trees under incomplete lineage sorting (ILS). CASTER outperforms traditional concatenation and gene tree summary methods like ASTRAL, SVDQuartets, and RAxML, particularly in high-ILS conditions, and scales to datasets with hundreds of species and billions of base pairs. The authors demonstrate its power by reconstructing the mammalian and avian species trees from whole genomes, identifying cases of introgression, recombination suppression, and alignment errors using per-site support scores. This work significantly advances genome-scale phylogenetics by enabling more accurate and comprehensive tree inference from entire genome alignments.
Methodological Highlights:
Site-based quartet weighting system: Assigns positive, negative, or zero weights to site patterns to handle ILS and long-branch attraction robustly.
Linear-time greedy tree search: Efficient algorithm to optimize quartet scores without enumerating all quartets, scaling to hundreds of species and billions of sites.
Per-site support scores: Provides site-level support for tree topologies, enabling the detection of introgression, recombination suppression, and alignment errors.
New Tools, Data, and Resources:
Code: Available at https://github.com/chaoszhang/ASTER. Written in C++, AGPL license.

scGPT-spatial: Continual Pretraining of Single-Cell Foundation Model for Spatial Transcriptomics
Paper: Wang, et al. "scGPT-spatial: Continual Pretraining of Single-Cell Foundation Model for Spatial Transcriptomics," bioRxiv, 2025. https://doi.org/10.1101/2025.02.05.636714.
Spatial transcriptomics complex. You need to model single-cell profiles while capturing spatial relationships all while handling diverse sequencing protocols (imaging-based vs. sequencing-based). scGPT-spatial is a foundation model built on the scGPT framework with MoE (mixture of experts) and pretrained on huge corpus of spatial transcriptomic profiles across technologies.
TL;DR: This paper introduces scGPT-spatial, a foundation model built to handle spatial transcriptomics data, extending the single-cell RNA-seq model scGPT. It integrates multi-modal spatial data and outperforms existing methods in key challenges in spatial omics like cell-type deconvolution and gene expression imputation.
Summary: scGPT-spatial is a continual pretrained model designed to bridge the gap between traditional scRNA-seq models and the spatial complexities inherent in spatial transcriptomics. The authors curated a massive dataset, SpatialHuman30M (30 million spatial transcriptomic profiles from platforms like Visium, Visium HD, MERFISH, and Xenium), to train the model. Two major innovations are highlighted: a Mixture-of-Experts (MoE) decoder that adapts to different sequencing protocols and a spatially-aware sampling strategy that captures local tissue microenvironments. scGPT-spatial showed superior performance in multi-modal integration, spatial domain clustering, cell-type deconvolution, and missing gene expression imputation, outperforming tools like Seurat, Tangram, and Cell2location without requiring additional fine-tuning. This model provides a robust, flexible framework for spatial omics analyses across diverse biological contexts.
Methodological Highlights (from senior author Bo Wang’s Tweet summary):
A Spatial-omic Foundation Model with Continual Pretraining – Built on scGPT’s robust initialization, it unlocks spatial context in tissues.
SpatialHuman30M Dataset – The largest curated dataset: 30M profiles from Visium, Visium HD, Xenium, and MERFISH across 821 slides.
Revolutionary MoE Decoders – A cutting-edge Mixture of Experts (MoE) architecture for protocol-aware gene expression decoding.
Spatially-Aware Training Strategy – A neighborhood-based masked reconstruction approach to capture complex cell-type colocalization.
Multi-Modal & Multi-Slide Integration – Seamless clustering & spatial domain identification across slides and modalities.
Cell-Type Deconvolution & Gene Imputation – Unlocks cross-resolution & cross-modality harmonization with fine-tuned embeddings.
New Tools, Data, and Resources:
scGPT-spatial code: Available at https://github.com/bowang-lab/scGPT-spatial (Python, MIT license). This repository includes the model code and instructions for downstream tasks like cell-type deconvolution and gene imputation.
Model weight are at FigShare: https://doi.org/10.6084/m9.figshare.28356068.v1.

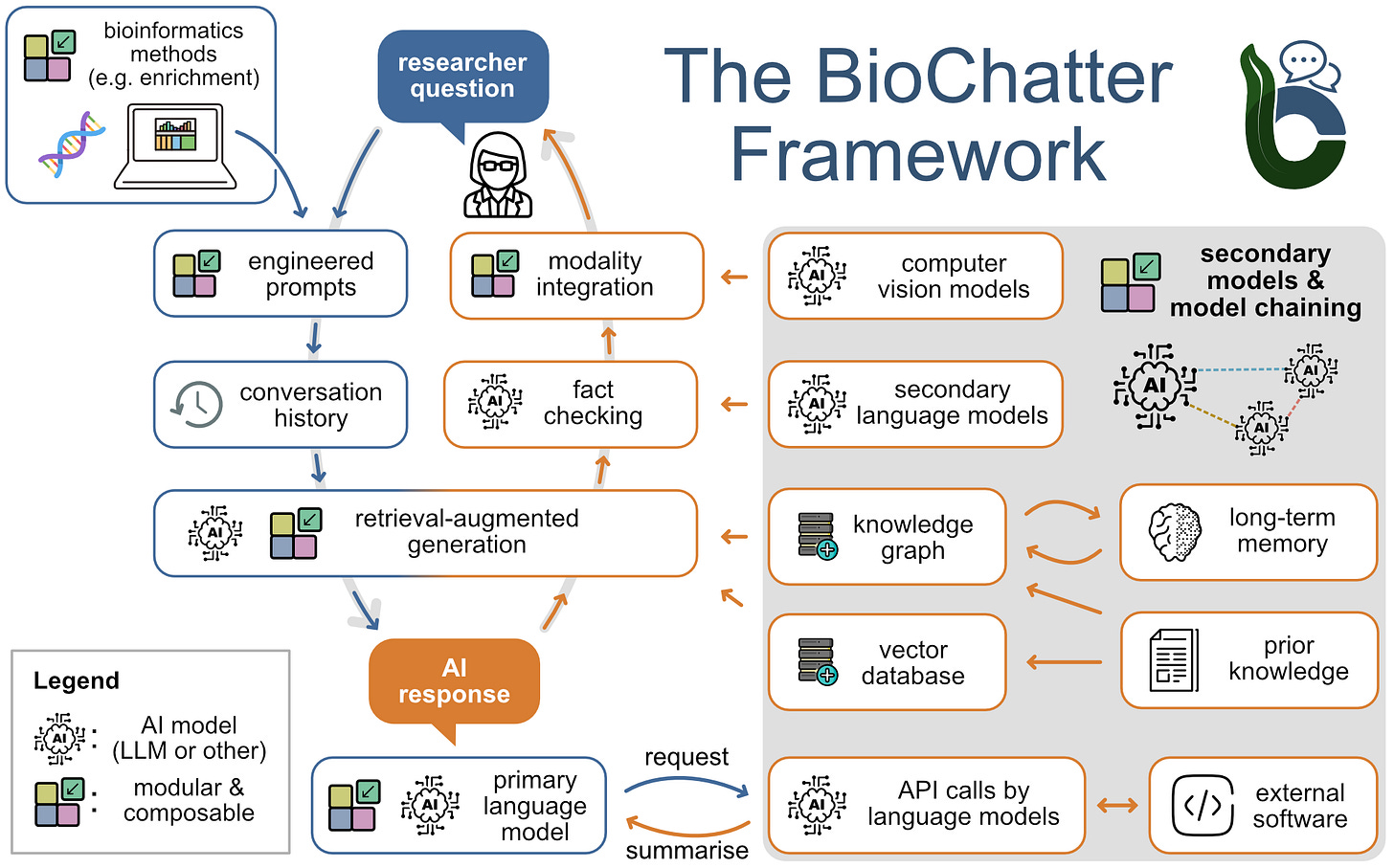
A platform for the biomedical application of large language models
Paper: Lobentanzer, et al. "A Platform for the Biomedical Application of Large Language Models," Nature Biotechnology, 2024. https://doi.org/10.1038/s41587-024-02534-3.
TL;DR: This paper introduces BioChatter, an open-source platform designed to bridge the gap between general-purpose large language models (LLMs) and their specific applications in biomedical research. It’s a Python package implementing a library for connecting biomedical applications to conversational AI.
Summary: BioChatter is a modular, open-source Python framework tailored for biomedical research applications using LLMs. The platform allows seamless integration with external tools (like OncoKB, BLAST) and knowledge graphs (e.g., BioCypher) to support tasks such as data extraction, hypothesis generation, and literature mining. It features flexible APIs, customizable system prompts, and multi-agent workflows for advanced data manipulation. BioChatter’s continuous benchmarking system ensures reproducibility and tracks model performance across different biomedical use cases. It improves knowledge graph query generation accuracy compared to naive LLM approaches. The platform aims to democratize LLM access in biomedicine, fostering community-driven development and open science practices.
Methodological Highlights:
Modular Architecture: Supports REST and Python APIs, with lightweight and advanced GUI options for flexible deployment in biomedical workflows.
Integration with Knowledge Graphs: Native support for BioCypher enables effective retrieval-augmented generation, enhancing data harmonization and model reliability.
Benchmarking Framework: Continuous performance tracking across models, with community-driven test cases to ensure robust, reproducible results.
Code: Available at https://github.com/biocypher/biochatter. Facilitates custom LLM-driven biomedical applications with multi-agent systems and API integration.
Mapping cells through time and space with moscot
Paper: Klein, et al. "Mapping Cells Through Time and Space with moscot," Nature, 2025. https://doi.org/10.1038/s41586-024-08453-2.
TL;DR: This paper introduces moscot, a robust optimal transport (OT) framework for mapping cells across time and space in single-cell multiomics and spatial transcriptomics. It scales OT methods to massive single-cell datasets (like 1.7M cells), works with multimodal data (e.g., gene expression + chromatin), and aligns developmental trajectories + spatial tissue sections.
Summary: moscot is a scalable OT framework built to address key challenges in single-cell and spatial transcriptomics like mapping cells across developmental time, aligning spatial sections, and integrating multimodal data. It introduces a unified API supporting Wasserstein (W), Gromov-Wasserstein (GW), and fused GW (FGW) OT to handle different biological data alignment tasks. The authors show it can reconstruct mouse embryogenesis trajectories from 1.7 million cells, map single-cell RNA + protein profiles to spatial liver transcriptomics, and align large spatial sections of the mouse brain. A standout result is the spatiotemporal mapping of mouse pancreas development using paired snRNA + ATAC data, revealing NEUROD2 as a regulator of epsilon cell differentiation (validated experimentally in human iPSC-derived islet cells).
Methodological Highlights:
Multimodal OT (RNA, ATAC, protein): Supports fused GW OT to align cells across different modalities (gene expression, chromatin, protein) and integrate with spatial data.
Low-rank Sinkhorn & OTT Library: Implements low-rank OT approximations and GPU-accelerated Sinkhorn algorithms (via JAX) to scale to atlas-level datasets with millions of cells.
Spatiotemporal Mapping: Combines temporal lineage mapping with spatial context (FGW) to trace cell fate during mouse development, enabling downstream analysis with CellRank.
New Tools, Data, and Resources:
Code: https://github.com/theislab/moscot (Python, BSD-3 license).
Documentation: https://moscot.readthedocs.io/
Mouse Embryogenesis Atlas (1.7M cells): Used to benchmark moscot for temporal lineage reconstruction; Available at http://tome.gs.washington.edu.

Reviews
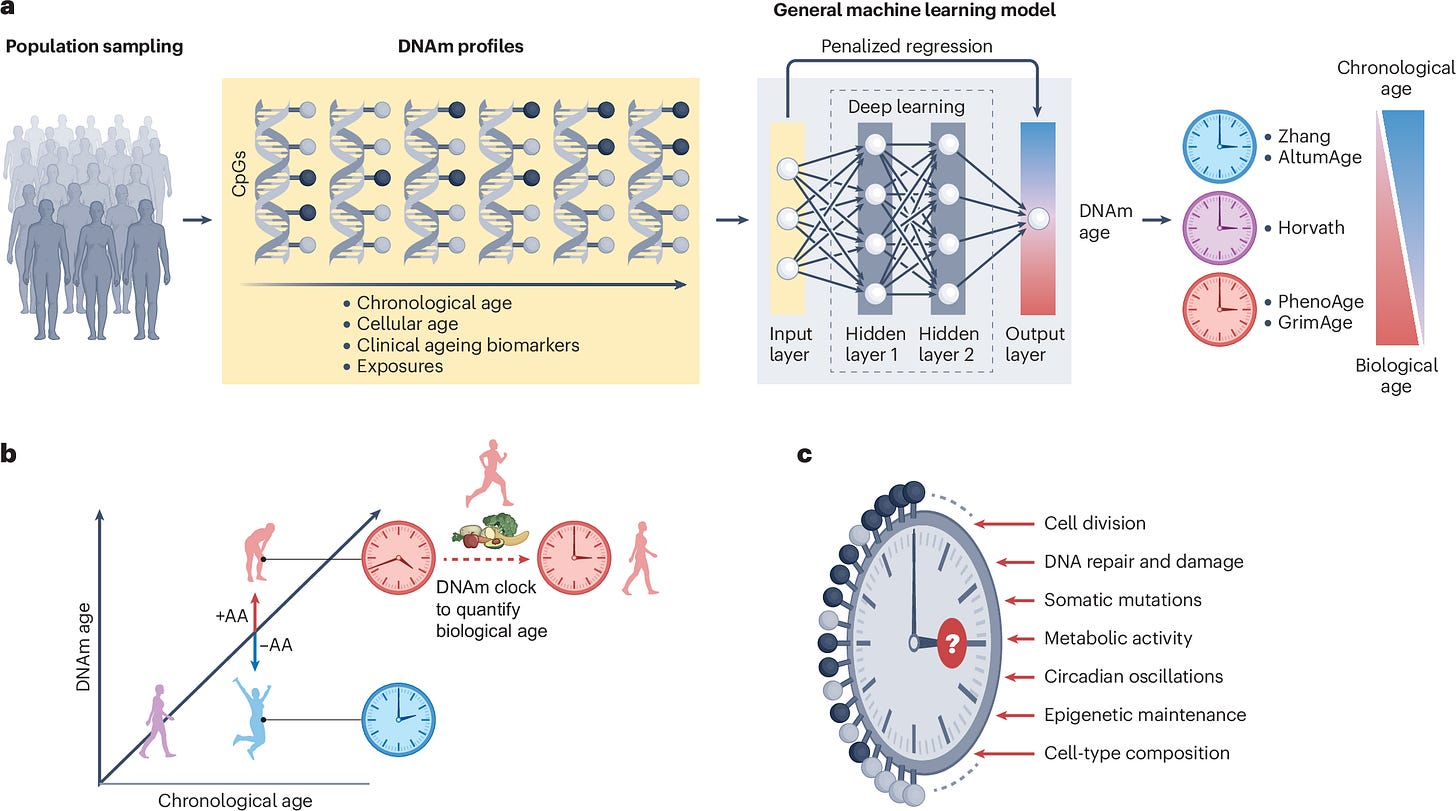
Epigenetic ageing clocks: statistical methods and emerging computational challenges
Paper: Teschendorff, A.E., & Horvath, S., “Epigenetic Ageing Clocks: Statistical Methods and Emerging Computational Challenges,” Nature Reviews Genetics, 2024. https://doi.org/10.1038/s41576-024-00807-w (read free: https://rdcu.be/d56ye).
This review paper breaks down the computational and statistical methods behind DNA methylation-based ageing clocks and highlights the major challenges around cell-type heterogeneity, single-cell approaches, and mechanistic interpretation. Over the past decade, DNA methylation (DNAm) clocks have revolutionized ageing research by providing molecular estimates of both chronological and biological age. These clocks are powerful tools for studying ageing processes, predicting health outcomes, and evaluating rejuvenation interventions. However, as the field advances, key computational and statistical challenges remain. This review dissects the underlying methods used to build DNAm clocks, such as elastic net regression and deep learning, while emphasizing emerging issues like cell-type heterogeneity and single-cell epigenetics. It also explores the biological interpretation of clock CpGs, the stochastic vs. deterministic nature of methylation changes, and the development of mechanism-specific clocks (e.g., mitotic clocks and clocks based on causal loci). Future progress will likely hinge on integrating single-cell data, building cell-type-specific clocks, and leveraging explainable AI to uncover the biological drivers of epigenetic ageing.
Diversity and Consequences of Structural Variation in the Human Genome
Paper: Collins, R.L., & Talkowski, M.E., "Diversity and Consequences of Structural Variation in the Human Genome," Nature Reviews Genetics, 2024. https://doi.org/10.1038/s41576-024-00808-9 (read free: https://rdcu.be/d9IEC).
This is a comprehensive review paper on structural variants (SVs) in the human genome, covering their mutational diversity, population genetics, functional impacts, and roles in human disease. Structural variants (genomic alterations ≥50bp) represent a diverse and functionally significant class of mutations, including deletions, duplications, inversions, insertions, and complex rearrangements. This review details the mutational mechanisms driving SVs, their prevalence across human populations, and their outsized impact on nucleotide diversity compared to single-nucleotide variants. The authors emphasize how long-read sequencing is expanding our understanding of SVs, revealing thousands of previously undetected variants in repetitive regions and showing that SVs often exert stronger effects on gene expression than smaller variants. SVs are under strong purifying selection due to their potential to disrupt gene dosage and regulatory architecture, yet specific cases of positive selection (e.g., AMY1 copy number variation) highlight their role in human adaptation. The review also covers SVs’ contributions to rare developmental disorders, common disease risk, and their growing importance in clinical diagnostics, while emphasizing future challenges in achieving accurate SV detection in complex genomic regions and integrating SVs into GWAS and functional studies.

Other papers of note
Massively parallel characterization of transcriptional regulatory elements https://www.nature.com/articles/s41586-024-08430-9
UDA-seq: universal droplet microfluidics-based combinatorial indexing for massive-scale multimodal single-cell sequencing https://www.nature.com/articles/s41592-024-02586-y
What large language models know and what people think they know https://www.nature.com/articles/s42256-024-00976-7
Challenges in detecting ecological interactions using sedimentary ancient DNA data https://www.biorxiv.org/content/10.1101/2024.08.16.608343v2
Overcoming challenges associated with broad sharing of human genomic data https://www.nature.com/articles/s41588-024-02049-2 (read free: https://rdcu.be/d68Kg)
Multiscale footprints reveal the organization of cis-regulatory elements https://www.nature.com/articles/s41586-024-08443-4
Integration of 168,000 samples reveals global patterns of the human gut microbiome https://www.cell.com/cell/fulltext/S0092-8674(24)01430-2
A genome-wide atlas of human cell morphology https://www.nature.com/articles/s41592-024-02537-7
Global meta-analysis shows action is needed to halt genetic diversity loss https://www.nature.com/articles/s41586-024-08458-x
A rapid chemical reprogramming system to generate human pluripotent stem cells https://www.nature.com/articles/s41589-024-01799-8 (read free: https://rdcu.be/d6bo3)

