Weekly Recap (December 19, 2025)
Writing better code without AI, AI in peer review, local LLMs, Biothreat Benchmark Generation, R updates (R Data Scientist, RWeekly, R Works), red-teaming an AI vending machine, new papers
Maëlle Salmon: Better Code, Without Any Effort, Without Even AI. lintr for detecting lints; Air for formatting code; jarl for detecting+fixing lints; flir for refactoring.
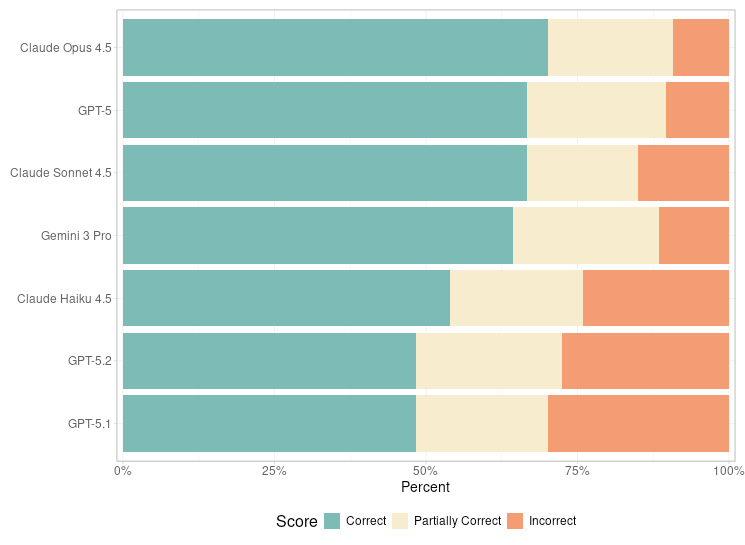
Update: Which LLM writes the best R code? Interestingly, GPT-5.2 underperformed OpenAI’s own GPT-5, as well as Gemini 3 and all the Claude models.
The R Data Scientist, 2025-12-17: Open Science & Pharma, Bioconductor, Genomics, Modeling Packages, LLMs for R, R, Dev Quality, DuckDB Pipelines, Visualization Storytelling, Spatial Analysis in R, Statistical Inference, Academic Research.
Nature News: More than half of researchers now use AI for peer review. Is anyone surprised? I imagine the number is actually higher than reported. Related, I wrote about prejudicial peer review with AI back over the summer:
Tim Dettmers: Why AGI Will Not Happen.
rOpenSci News Digest, December 2025.
Simon Couch: chores 0.3.0 and local LLMs. TL;DR:
The chores package, up to this point, needed a frontier-ish model to be useful; local models were more trouble than they were worth.
Qwen3 4B Instruct 2507 is good enough, and it’s small enough to comfortably run on high-end laptops.
Simon Willison: Useful patterns for building HTML tools with AI.
Simon Willison, again: Your job is to deliver code you have proven to work.
As software engineers we don’t just crank out code—in fact these days you could argue that’s what the LLMs are for. We need to deliver code that works—and we need to include proof that it works as well. Not doing that directly shifts the burden of the actual work to whoever is expected to review our code.
Adam Kucharski: When is the ‘ground truth’ not quite the whole truth?
Related, Adam Kucharski has started writing a weekly recap on his newsletter, Understanding the Unseen (which I recommend reading):
Johann Rehberger: The Normalization of Deviance in AI.
Nature news article: Huge genetic study reveals hidden links between psychiatric conditions. Analysis of more than 1 million people shows that mental-health disorders fall into five clusters, each of them linked to a specific set of genetic variants. See the original paper in Nature: Mapping the genetic landscape across 14 psychiatric disorders.
The US OPM+OMB+OSTP+GSA lanch the U.S. Tech Force: techforce.gov.
R Weekly 2025-W51: data visualisation, assess usage of your package, mutagen.
Science news story on aiXiv (https://aixiv.science/) — a new preprint server that welcomes papers written and reviewed by AI. “aiXiv is a free, AI- or human-peer-reviewed preprint archive for research authored by AI Scientists, Robot Scientists, Human Scientists across all scientific fields. We believe that groundbreaking research should be accessible to everyone, everywhere. Our platform harnesses the power of artificial intelligence and robotics to accelerate scientific discovery, enhance collaboration, and democratize knowledge sharing.” I’m generally “pro-AI-of-Center” but a preprint server with AI reviews of papers written by AI that no one will ever actually read is most dystopic waste of time and resources I’ve heard about in a while. The few AI authored papers I clicked through had a “corresponding human” with affiliations listed as “independent scientist” using a throwaway Gmail address, who very well could be fake people. I’m actually a little surprised Science spent the time covering something so silly.
Biothreat Benchmark Generation Framework for Evaluating Frontier AI Models:
Maëlle Salmon @ rOpenSci: How to Assess Usage of your Package. I learned an interesting trick here using GitHub’s search. As some code is published on GitHub, you can use advanced GitHub searches to find instances of use. You can search for occurrences of, say, library(qqman) using the URL https://github.com/search?q=library(qqman)&type=code. Interesting to see this package that I haven’t touched in nearly a decade being used in nearly 3,000 files. Especially since there are much better alternatives out there.
Science / Science Insider: NIH’s proposed caps on open-access publishing fees roil scientific community.
NSF Merit Review For a Changing Landscape (PDF).
WSJ let Claude run a vending machine in their office. It ordered a live fish. It offered to buy stun guns, pepper spray, cigarettes and underwear. Profits collapsed. Newsroom morale soared.
Finally, a few other papers and preprints that caught my attention this week:
Enzyme Engineering Database: a platform for sharing and interpreting sequence–function relationships across protein engineering campaigns (enzengdb.org)
Pyranges v1: a Python framework for ultrafast sequence interval operations
Scikit-bio: a fundamental Python library for biological omic data analysis
Bioinformatics frameworks for single-cell long-read sequencing: unlocking isoform-level resolution
dtreg: Describing Data Analysis in Machine-Readable Format in Python and R