Gene Info Custom GPT
Gene Info: a custom GPT that takes a list of gene symbols and provides summary information, gene ontology terms, and provides contextual information like pathway or disease involvement.
OpenAI introduced the ability to create custom GPTs back in November 2023. I wanted to try to create one of these, and in the spirit of learning in public this post describes how I made it. But first, what does it do?
Gene Info custom GPT
The Gene Info custom GPT takes a list of human gene symbols as input. It'll run some Python code against a custom knowledge base to provide information about those genes (from RefSeq).
Here’s the start page interface. The example chat starters in the GPT are genes known to be involved in (1) apoptosis, (2) cell differentiation, (3) innate immunity, and (4) RNA processing.
Let’s try set number 3 (genes involved in innate immunity). The first thing you’ll see returned is a table with your inputs linked to information about those genes.
If you click the little [>_] symbol at the end, it’ll show you the code it used to create this table.
You can click on the expand arrow ↔️ in the table to expand those results to a full screen view. You can also hit the download arrow ⬇ to download a CSV with these results.
The GPT will suggest some follow up questions. What about pathways these genes are involved in?
Or what about what diseases these genes are involved in?
Give it a try now if you’re interested.
How I built it
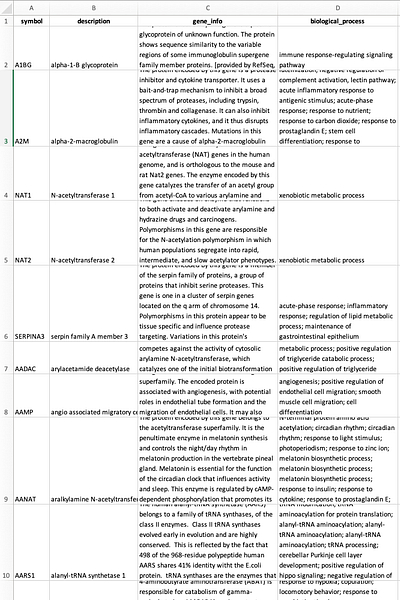
“How I built it” is generous here. I really didn’t do much “building.” First, I grabbed gene summary info for all genes, human gene information, and gene to gene ontology term tables from the RefSeq FTP site, joined them all together and pulled out only the columns I care about, and wrote the resulting table as CSV, XLSX (for uploading here), and Parquet. The R code to do this is here as a GitHub Gist. Here’s that table.
I then provided that table as uploaded knowledge, together with a system prompt telling the custom GPT what to do. I seeded it with starter conversations containing lists of 20 genes, each known to be involved in (1) apoptosis, (2) cell differentiation, (3) innate immunity, and (4) RNA processing, respectively. I gave it access to code interpreter, web search, and canvas. I published it for anyone with the link.

Coda
Admittedly there are some shortcomings here. The GPT doesn’t have access to actual biological pathway information (only gene ontology biological processes), nor does it have access to disease associations (these are probably from the base GPT-4o model). A future iteration of this tool could actually retrieve things like KEGG pathways, GWAS catalog associations, OMIM data, or similar.
This was a 10 minute hobby project I created just to learn how to create a custom GPT. There are others putting actual effort into things like this (e.g., CurateGPT as one example upon many). What I’d really like to do is create an actual RAG app, self-hosted, built using Llama3.3-70B running via open-webui. That’s a future post.