Weekly Recap (Oct 2024, part 2)
Gene-level alignment of scRNA-seq trajectories, integrating identifiers across databases, SVs in humans and apes, universal prediction of cellular phenotypes, and a bioinformatics analysis chatbot
This week’s recap highlights a new method for gene-level alignment of single-cell trajectories, an R package for integrating gene and protein identifiers across biological sequence databases, characterization of SVs across humans and apes, universal prediction of cellular phenotypes, a method to quantify cell state heritability versus plasticity and infer cell state transition with single cell data, and a new AI-driven, natural language-oriented bioinformatics pipeline assists with automatic and codeless execution of biological analyses.
Others that caught my attention include pangenome-informed privacy preserving synthetic sequence generation, a paper showing generative haplotype prediction outperforms statistical methods for small variant detection, a metadata standardizer for genomic region attributes, a web-based platform for reference-based analysis of single-cell datasets, a new taxa-specific normalization approach for microbiome data, a deep-learning-based splice site predictor, models for human metabologenomics, a Snakemake pipeline to automatically generate pangenomes from metagenome assembled genomes and a new paper on COVID-19 origins.
Deep dive
Gene-level alignment of single-cell trajectories (Genes2Genes)
Paper: Sumanaweera et al., "Gene-level alignment of single-cell trajectories," Nature Methods, 2024. DOI: 10.1038/s41592-024-02378-4.
Genes2Genes appears to offer a new way to accurately match and compare single-cell trajectories at the gene level, helping to spot differences that other methods miss.
TL;DR: This paper introduces Genes2Genes (G2G), a Bayesian dynamic programming framework for aligning single-cell trajectories at the gene level. It overcomes limitations of traditional dynamic time warping (DTW) by accurately capturing sequential matches and mismatches, providing precise trajectory alignments in various biological contexts, including disease cell-state analysis.
Summary: The study presents Genes2Genes (G2G), a novel framework for aligning single-cell pseudotime trajectories at the resolution of individual genes. Traditional approaches like dynamic time warping (DTW) often assume that each time point in a reference trajectory has a corresponding point in the query trajectory, and they cannot adequately handle mismatches like insertions or deletions. G2G addresses these limitations by extending DTW with a Bayesian information-theoretic framework, implementing a dynamic programming (DP) algorithm that identifies five alignment states: match, expansion warp, compression warp, insertion, and deletion. The method uses minimum message length (MML) inference to compute the optimal alignments, capturing both matches and mismatches in gene expression profiles. Experiments on simulated datasets and real-world single-cell RNA-seq data demonstrate G2G's superior performance in aligning trajectories and identifying clusters of gene alignment patterns. For instance, G2G accurately aligns T cell differentiation trajectories in vitro with those in vivo, revealing that in vitro differentiation misses key gene expression patterns associated with TNF signaling, guiding the optimization of culture conditions to better recapitulate in vivo states. This method holds potential for enhancing our understanding of dynamic cellular processes and refining therapeutic interventions.
Methodological highlights:
Introduces a dynamic programming (DP) algorithm that combines DTW with Bayesian scoring to capture detailed alignment patterns, including mismatches and insertions/deletions, at single-gene resolution.
Uses a five-state model (match, expansion warp, compression warp, insertion, deletion) to align time points between reference and query trajectories, allowing for more flexible and accurate comparisons.
Applies minimum message length (MML) inference for scoring, accounting for both mean and variance differences in gene expression distributions.
New tools, data, and resources:
Genes2Genes (G2G) software: G2G is an open-source Python package with a tutorial available at https://github.com/Teichlab/Genes2Genes. Code used to perform analyses in the manuscript using G2G v.0.1.0 is available at https://github.com/Teichlab/G2G_notebooks.
Data availability: Data used to perform analyses in the manuscript are available at https://zenodo.org/records/11182400 and https://github.com/Teichlab/G2G_notebooks. All generated alignments are available as Supplementary Data. Raw sequencing data for newly generated sequencing libraries are in ArrayExpress (accession no. E-MTAB-12720).

ginmappeR: an unified approach for integrating gene and protein identifiers across biological sequence databases
Paper: Sola et al., "ginmappeR: an unified approach for integrating gene and protein identifiers across biological sequence databases," Bioinformatics Advances, 2024. DOI: doi.org/10.1093/bioadv/vbae129.
Dealing with gene and protein names across databases such as GenBank, UniProt, KEGG, etc. is the up there along with 0/1 open/closed interval formats as a candidate for the most eternally frustrating parts of bioinformatics. This R package looks promising as a tool to help with gene ID mapping.
TL;DR: The ginmappeR R package facilitates the translation of gene and protein identifiers between major biological sequence databases, improving data integration for bioinformatics workflows. It provides a unified interface and enhances usability through features like caching and error handling.
Summary: This paper introduces ginmappeR, an R package designed to address the challenges of integrating gene and protein identifiers across various biological sequence databases, such as NCBI, UniProt, KEGG, and CARD. With the growing volume of biological data, inconsistencies in identifiers pose significant challenges for researchers. ginmappeR offers a unified interface for translating these identifiers, simplifying integration into bioinformatics workflows and reducing manual efforts. It also features caching and robust error handling mechanisms to enhance efficiency and reliability. This package is particularly useful for studies involving multiple data sources and large-scale analyses, enabling seamless integration of diverse data and improving workflow automation.
Methodological highlights:
Consistent interface: Provides functions for mapping gene/protein identifiers between multiple databases using a consistent format.
Enhanced reliability: Implements caching and error handling layers to prevent redundant API calls and manage API failures.
On-the-fly translations: Ensures users work with the most current data available by querying databases in real time, unlike tools that rely on periodically updated backends.
ginmappeR package: The package is available on Bioconductor at https://bioconductor.org/packages/ginmappeR. The source code is on GitHub at https://github.com/DEAL-US/ginmappeR.
Data sources: Supports NCBI, UniProt, KEGG, and CARD databases. Uses public APIs or local files for data retrieval.

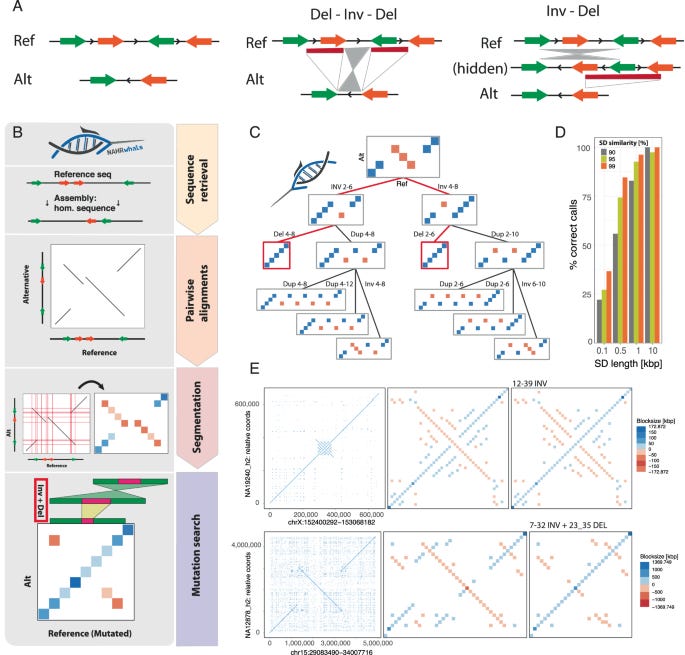
Impact and characterization of serial structural variations across humans and great apes
Paper: Höps et al., "Impact and characterization of serial structural variations across humans and great apes," Nature Communications, 2024: 10.1038/s41467-024-52027-9.
The NAHRwhals method introduced here finds overlapping structural variation that occur because of non-allelic homologous recombination (NAHR). It’s an R package.
TL;DR: This paper presents NAHRwhals (NAHR-directed Workflow for catcHing seriAL Structural Variations), a new computational tool for identifying complex serial structural variations (sSVs) in genomes using long-read assemblies. The study explores these variations in human and great ape genomes, revealing their potential role in genetic diversity and disease.
Summary: The study introduces NAHRwhals, a method developed to detect serial structural variations (sSVs), which are complex DNA rearrangements caused by repeated mutation events. Unlike traditional SV detection tools, NAHRwhals identifies series of overlapping SVs that occur due to non-allelic homologous recombination (NAHR) or other mechanisms. Applied to human and great ape genomes, NAHRwhals uncovered 37 sSV loci, highlighting their importance in evolutionary biology and medical genetics. These sSVs often occur in regions with high genetic diversity and have implications for understanding disease susceptibility and human evolution. For instance, sSVs found in the TPSAB1 gene region and others could explain cryptic variation related to diseases like Sotos syndrome. The tool can be applied to any species, making it valuable for broad genomic studies.
Methodological highlights:
NAHRwhals is an R package providing tools for visualization and automatic detection of complex, NAHR-driven rearrangements (few kbp to multiple Mbp) using genome assemblies. Modules include:
Liftover of coordinates between arbitrary human DNA assemblies
Accurate sequence alignments of multi-MB DNA sequences
Dotplot visualizations
Segmented Dotplots
A tree-based caller for complex, nested NAHR-mediated rearrangements.
Exhaustive search algorithm: NAHRwhals employs a depth-first search to identify sSVs up to a predefined depth, enabling it to map complex genetic rearrangements.
High-resolution pairwise alignments: The tool uses a custom alignment strategy to accurately map repetitive genomic regions, improving detection in areas where traditional methods struggle.
Genotyping and whole-genome modes: NAHRwhals can focus on specific genomic regions or scan entire genomes for sSVs, offering flexibility in its applications.
New tools, data, and resources:
Code availability:
NAHRwhals is available on github under an MIT license: https://github.com/WHops/NAHRwhals.
Code for SV phasing and integration with external SNPs (population-based analysis) is available at https://github.com/WHops/nahrwhals_phasing.
Visualizations of Strand-Seq data were made with a custom pipeline available at: https://github.com/WHops/sseq_plot.
Code used to create artificial sequences and their mutated counterparts for simulation-based benchmarking is available at https://github.com/WHops/nahrwhals_simulate_events.
Data availability: The data availability section of the paper provides links to data available from HGSVC, IGSR, Zenodo, and other sources.
Scalable and universal prediction of cellular phenotypes
Paper: Ji et al., "Prophet: Scalable and universal prediction of cellular phenotypes," bioRxiv, 2024. https://doi.org/10.1101/2024.08.12.607533
So much of life sciences research from biomedicine to de-extinction involves modeling or prediction of phenotype from “exposure” to a genotype, drug, genetic perturbation, etc. The prophet tool introduced here predicts cellular phenotypes (expression, viability, etc) of untested small molecule or genetic perturbations.
TL;DR: Prophet is a transformer-based model for predicting cellular phenotypes across various experimental conditions. It uses transfer learning to generalize across diverse datasets, making it a powerful tool for guiding biological experimentation and drug discovery.
Summary: The study introduces Prophet, a novel transformer-based model designed to predict cellular phenotypes like gene expression and cell viability across a wide range of experimental conditions, including different cell types and perturbations (e.g., drugs, genetic modifications). Prophet employs a three-axis framework that includes cellular state, treatment, and phenotypic readout, enabling it to learn from and integrate data across nine large-scale perturbation datasets. The model leverages transfer learning to predict unseen experimental outcomes with high accuracy and can suggest promising experimental conditions to explore, effectively reducing the need for extensive experimental screening. Prophet’s scalability and robust performance position it as a versatile tool for accelerating biological discovery and experimental design.
Methodological highlights:
Transformer-based architecture: Utilizes 8 transformer encoder units and 8 attention heads per layer, enabling effective learning of complex relationships between cell states, treatments, and phenotypes.
Transfer learning: Pretrained on 4.7 million experiments and fine-tuned for specific datasets, demonstrating strong performance across unseen conditions.
Set-based representation: Models experiments as sets of tokens representing cellular states, treatments, and readouts, allowing flexible predictions across diverse phenotypic spaces.
New tools, data, and resources:
Prophet model: Available on GitHub at https://github.com/HelmholtzAI/Prophet (note the noncommercial license).
Large perturbation dataset: Data obtained from in vitro validation can be found at https://figshare.com/articles/dataset/Prophet_viability_plate_xlsx/26516002.

Defining heritability, plasticity, and transition dynamics of cellular phenotypes in somatic evolution
Paper: Schiffman et al., "Defining heritability, plasticity, and transition dynamics of cellular phenotypes in somatic evolution," Nature Genetics, 2024. DOI: 10.1038/s41588-024-01920-6. Read free: https://rdcu.be/dUWvn.
PATH is an R package for analyzing multi-modal single-cell phylogenies.
TL;DR: The study introduces PATH (phylogenetic analysis of trait heritability), a framework for quantifying the heritability and plasticity of cellular phenotypes using single-cell lineage tracing data. Applied to cancer models and human glioblastoma samples, PATH reveals complex dynamics of cell state transitions and the implications for tumor evolution and therapy.
Summary: This study presents PATH, a phylogenetic framework that quantitatively measures the heritability and plasticity of cellular phenotypes and infers cell state transitions using single-cell lineage tracing data. PATH applies phylogenetic correlations to model the relationship between cell states and their ancestral lineages, providing a metric for phenotypic heritability. The tool is tested on several models, including a mouse model of pancreatic cancer and primary human glioblastoma samples, demonstrating its ability to map complex cellular dynamics. In glioblastoma, PATH identified bidirectional transitions between stem-like and mesenchymal-like states, using an astrocyte-like state as an intermediary. The findings highlight how varying plasticity across cell states can influence cancer progression and therapeutic resistance, making PATH a valuable tool for dissecting the evolutionary dynamics of tumors and other somatic tissues.
Methodological highlights:
Phylogenetic correlation metric: Quantifies the heritability versus plasticity of cellular phenotypes by measuring how similar cell states are among related cells compared to random cells.
PATHpro extension: Incorporates cell state-specific proliferation rates into the model, improving transition and proliferation dynamics inferences even when proliferation rates vary significantly.
High computational efficiency: PATH offers faster computation compared to maximum likelihood estimation (MLE), making it suitable for analyzing large single-cell lineage datasets.
New tools, data, and resources:
PATH software: The PATH R package is available on GitHub and can be installed with devtools: https://github.com/landau-lab/PATH. This interactive PATH Simulator web app can also be used for simple demonstrations.
Data availability: Data used in this paper come from GEO: GSE173958, GSE151506, and GSE273357.

BioMANIA: Simplifying bioinformatics data analysis through conversation
Paper: Dong et al., "BioMANIA: Simplifying bioinformatics data analysis through conversation," bioRxiv, 2024. https://doi.org/10.1101/2023.10.29.564479.
There’s no lack of new LLM-based tools, agents, chatbots, etc. for assisting with analysis and interpretation of sequencing data and bioinformatics analysis. BioMANIA is another one, and it seems to be limited to Python tools only. An interesting, if not obvious claim, is that the effectiveness of a conversational data analysis is highly dependent on the design and implementation of the underlying (Python) tools that BioMANIA uses. A quote from the discussion section:
The Python tools used by BioMANIA exhibit varying levels of API design ambiguity, validity, and complexity, which are demonstrated to influence the effectiveness of conversational data analysis. These factors can lead to challenges such as ambiguous function names, excessive and poorly documented parameters, or out- dated documentation, all of which can hinder accurate API prediction, argument selection, and overall performance of the conversational interface. This limitation highlights an area for future development.
TL;DR: BioMANIA is an AI-driven tool that enables codeless, conversational bioinformatics data analysis. It leverages large language models to interpret natural language instructions and automate complex workflows across various bioinformatics tools, reducing technical barriers for researchers.
Summary: This paper introduces BioMANIA, a novel natural language-driven bioinformatics platform designed to simplify data analysis tasks for experimental researchers who may lack advanced programming skills. By integrating large language models (LLMs) with domain-specific APIs from existing bioinformatics tools, BioMANIA translates user instructions into executable code, automating complex workflows in areas like single-cell omics and electronic health record analysis. The tool provides a user-friendly interface, enabling researchers to perform sophisticated analyses without needing to write code. BioMANIA is benchmarked against traditional LLMs and general-purpose bioinformatics tools, demonstrating superior performance in executing and troubleshooting data analysis tasks. This approach significantly lowers the barrier to performing high-quality bioinformatics analyses, facilitating more effective and accessible research.
Methodological highlights:
Conversational pipeline: Uses a multi-step process to interpret user intent, map it to API calls, and execute those calls with inferred arguments, providing a structured and error-handling approach.
API integration and retriever: Integrates APIs from well-documented bioinformatics tools and employs a fine-tuned retriever to accurately map user instructions to relevant APIs.
Automatic troubleshooting: When errors occur, BioMANIA iteratively revises API calls based on error information, reducing the need for manual intervention.
New tools, data, and resources:
BioMANIA
Code is available at https://github.com/batmen-lab/BioMANIA.
Docker images: https://hub.docker.com/repositories/chatbotuibiomania
Benchmark datasets: The data used in this paper is available from a Google Drive link in the paper. I’m always wary of linking to Google Drive folders, as the contents can change. Take a look at the paper if you want the data.

Other papers of note
Pangenome-Informed Language Models for Privacy-Preserving Synthetic Genome Sequence Generation https://www.biorxiv.org/content/10.1101/2024.09.18.612131v1
Generative Haplotype Prediction Outperforms Statistical Methods for Small Variant Detection in NGS Data https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btae565/7762102
BEDMS: A metadata standardizer for genomic region attributes https://www.biorxiv.org/content/10.1101/2024.09.18.613791v1
archmap.bio: A web-based platform for reference-based analysis of single-cell datasets https://www.biorxiv.org/content/10.1101/2024.09.19.613883v1
Taxanorm: a novel taxa-specific normalization approach for microbiome data https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-024-05918-z
Splam: a deep-learning-based splice site predictor that improves spliced alignments https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03379-4
Genome-scale models in human metabologenomics https://www.nature.com/articles/s41576-024-00768-0 (read free: https://rdcu.be/dUsas)
CELEBRIMBOR: Core and accessory genes from metagenomes https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btae542/7762100
Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic https://www.cell.com/cell/fulltext/S0092-8674(24)00901-2
